Comprehensive user guide
Comprehensive tips for reliable and efficient analysis set-up
Objective
We have prepared this guide to help you with your first set of projects on BioData Catalyst powered by Seven Bridges. Each section has specific examples and instructions to demonstrate how to accomplish each step. We also highlight potential stumbling blocks so you can avoid them as you get set up. If you need more information on a particular subject, our Knowledge Center has additional information on all of the platform features. Additionally, our support team is available 24/7 to help!
Helpful terms to know
Tool refers to a stand-alone bioinformatics tool or its Common Workflow Language (CWL) wrapper that is created or already available on the Platform.
Workflow / Pipeline (interchangeably used) – denotes a number of tools connected together in order to perform multiple analysis steps in one run.
App stands for a CWL wrapper of a tool or a workflow that is created or already available on the platform.
Task – represents an execution of a particular tool or workflow on the platform. Depending on what is being executed (tool or workflow), a single task can consist of only one tool execution (tool case) or multiple executions (one or more per each tool in the workflow).
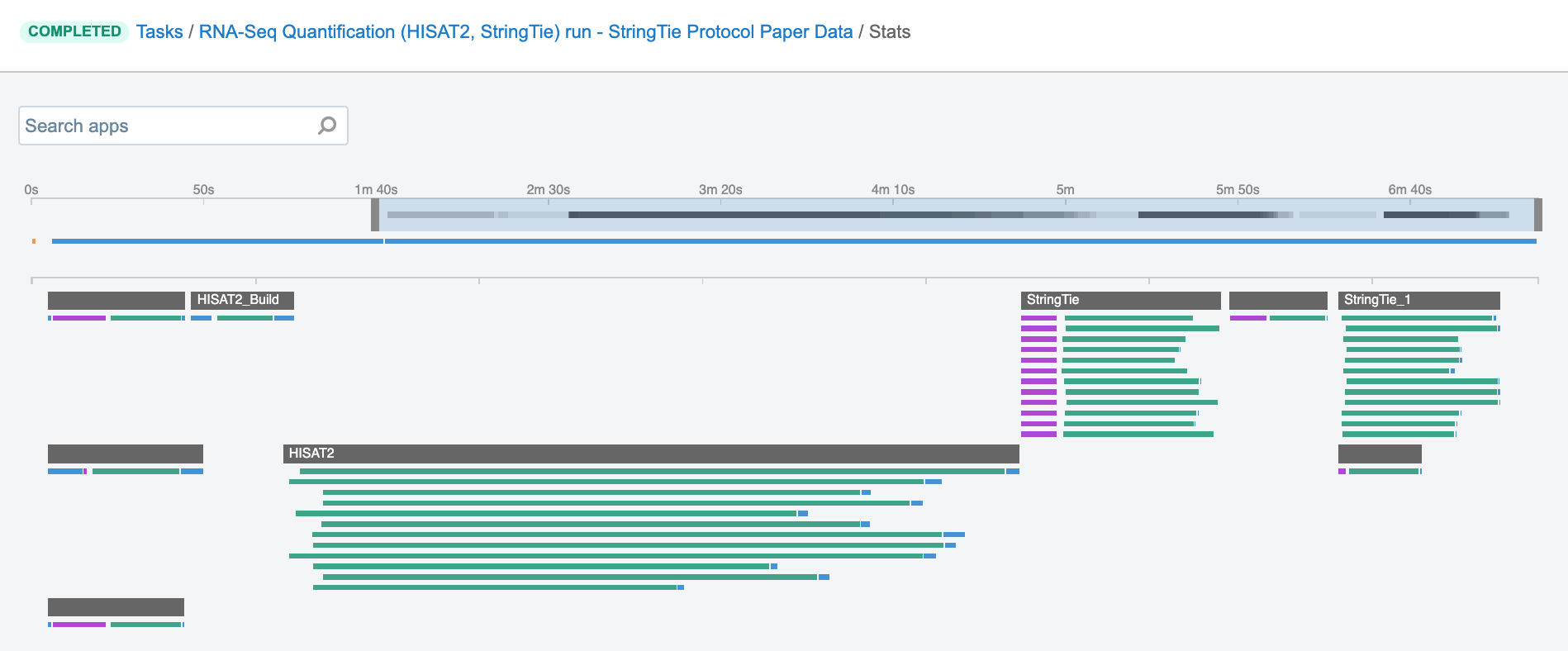
Job – this refers to the “execution” part from the “Task” definition (see above). It represents a single run of a single tool found within a workflow. If you are coming from a computer science background, you will notice that the definition is quite similar to a common understanding of the term “job” (wikipedia). Except that the “job” is a component of a bigger unit of work called a “task” and not the other way around, as in some other areas may be the case. To further illustrate what job means on the platform, we can visually inspect jobs after the task has been executed using the View stats & logs panel (button in the upper right corner on the task page):
Figure 1. The jobs for an example run of RNA-Seq Quantification (HISAT2, StringTie) public workflow.The green bars under the gray ones (apps) represent the jobs (Figure 1). As you can see, some apps (e.g. HISAT2_Build) consist of only one job, whereas others (e.g. HISAT2) contain multiple jobs that are executed simultaneously.
User Accounts & Billing Groups
To access BioData Catalyst powered by Seven Bridges, you need an eRA Commons account. We encourage researchers to register (instructions in the documentation) and discover numerous methodologies to analyze NHLBI datasets in a streamlined and efficient manner. If you would like to analyze the hosted controlled data on the platform, you will need approval for that data in dbGaP.
Upon registration, each user receives $500 in credits in a "Pilot Fund" billing group to get started. Billing groups track cloud costs (compute and storage) incurred by the user and each project on the platform must be assigned to a billing group. Seven Bridges will contact users when they are close to consuming all of their Pilot Fund credits to discuss getting a paid billing group set up. This billing group could use awarded cloud credits from NHLBI or a credit card or purchase order.
Work on the platform is divided into separate workspaces called projects (Figure 2). Each user can create projects and invite other registered users to be members of those projects. In addition, a variety of permission levels (e.g. admin, write, read-only) can be assigned to each project member.
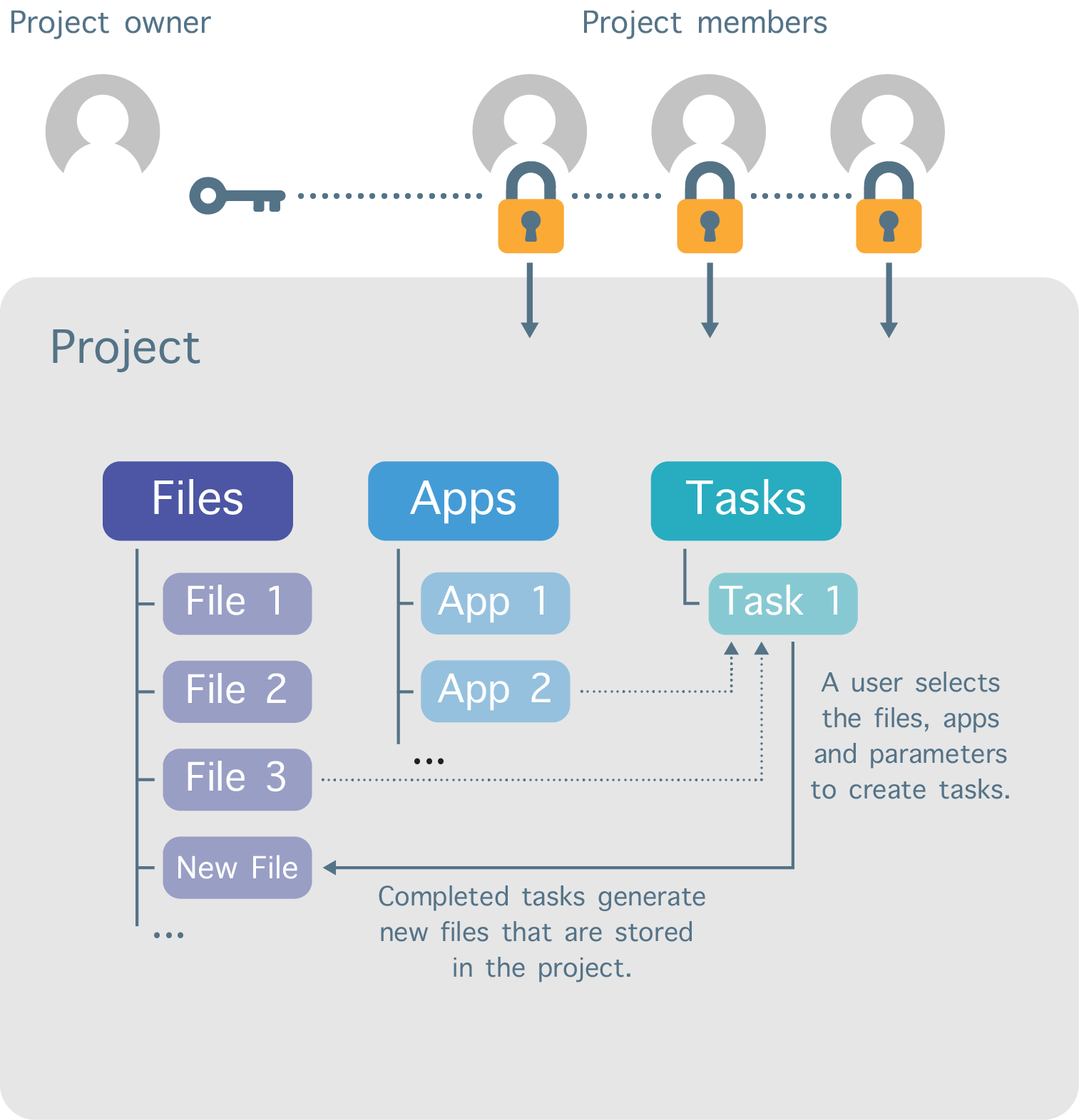
Figure 2. Project organization structure.
The user that creates the project sets the billing group to which the storage and compute costs are billed. The billing group for a project can be changed at any time.

Further Reading
Sign up for BioData Catalyst powered by Seven Bridges:
https://sb-biodatacatalyst.readme.io/docs/sign-up-biodata-catalyst-powered-by-seven-bridges
Before you start:
https://sb-biodatacatalyst.readme.io/docs/before-you-start
Access via the visual interface:
https://sb-biodatacatalyst.readme.io/docs/access-via-the-visual-interface
Projects on BioData Catalyst powered by Seven Bridges:
https://sb-biodatacatalyst.readme.io/docs/projects-on-the-platform
Add a collaborator to a project:
https://sb-biodatacatalyst.readme.io/docs/add-a-collaborator-to-a-project
Change the billing group for a project:
https://sb-biodatacatalyst.readme.io/docs/modify-project-settings
Account settings and billing:
https://sb-biodatacatalyst.readme.io/docs/account-settings-and-billing
Tips for Running Tools/Workflows
In this section you will find some of the most essential information on how to set up and run tools and workflows on the platform. Creating CWL wrappers is not in the scope of this section; instead, we focus on platform features which are fundamental to making your analysis efficient and scalable. The content is structured such that it first offers information that is helpful for those researchers who want to use the public tools and workflows, and then it gradually advances to describe the features which can be used to adjust the existing tools or develop new ones.
Start with the descriptions
For users who plan to execute one of the hundreds of hosted tools or workflows from the Public Apps Gallery, the most important step is to thoroughly read the description. Even if you are familiar with the tools or you regularly run similar tools on your local machine, the description is where you will find useful information on what you can expect in terms of the analysis design, expected analysis duration, expected cost, common issues, and more. The Common Issues and Important Notes section is an essential section in the description which gives insights into the known pitfalls you may come across when running the tool and should be studied the most, especially when planning to run large scale analysis.
Although Seven Bridges aims to improve the public apps and prevent users from using them incorrectly, there are many places for error which can cause the tools/workflows to not complete successfully when executed. Below is an example of how overlooking the description notes caused an unexpected outcome for a researcher:
Bioinformatics tools are usually designed to fail if certain inputs are configured incorrectly, and therefore terminate the misconfigured execution as soon as possible. However, in the case of VarDict variant caller, failing to provide the required input index files won’t always result in the failure, but can sometimes lead to infinite running.
This outcome occured when a user tried to process a number of samples by using the VarDict Single Sample Calling workflow. Previously, the user analyzed a whole cohort of samples with this workflow without any issues, and then tried to apply the same strategy to another dataset in a different project. When setting up the workflow in the new project, the user copied the reference files to the new project and initiated the workflow with this new batch of files. However, the user left out the reference genome index file (FAI file) during the copying step, in turn producing several tasks which suffered from infinite running. Since the index files are automatically loaded from the project and not referenced explicitly as separate inputs, it was even more likely for this issue to go unnoticed.
The situation was additionally compounded by having a batch task (see Batch Analysis section for more details) which contained hundreds of children tasks. Because the user wasn’t familiar with the related notes from the workflow description, they were not aware that the running tasks might in fact be hanging tasks. Had they known, they would have been able to terminate the run earlier, saving time and money.
In this case, the majority of the tasks failed early in the execution, producing minimal charges, but a few frozen tasks doubled the costs expected for the entire batch. We have since addressed the issues in this particular tool by introducing additional checks and input validations in the wrapper. Even though this is an extreme example, it demonstrates how descriptions can be critical to your tool working properly and can help you make informed decisions.
Test the workflow
If you are planning a large scale analysis, we recommend beginning by testing the workflow in a small number (1-5) of runs. By testing your tools on a small scale, you can quickly make sure that everything works as expected, while minimizing costs. Additionally, testing the pipeline for the worst case scenario is one of the best ways to get more insight into the potential edge cases and know what to expect. This can often be accomplished by running the analysis with the files of largest size in your dataset. There are other situations where the highest computational load will correlate with the complexity of the sample content, which is more difficult to estimate. Familiarity with the experimental design that went into your sample files helps to create proper test cases, for both size and complexity, which can keep potential errors to a minimum.
Specify computational resources
The term “computational instance” or “instance” refers to a virtual machine that can be chosen in order to adjust CPU and memory capacities for the particular application. The instances can be selected from the predefined list of Amazon EC2 or Google Cloud instance types.
For public apps, the instance resources have been pre-tested and defined by the Seven Bridges team. In most cases, the pre-defined instance types and default parameters will work for the majority of workflows, but you may need to optimize for certain input sizes or complexities, especially when scaling up your workflow.
Small-scale testing can help inform whether or not you need to tweak the instance type(s) or specific tool parameters you will use throughout the analysis. There are two means by which instance selection is controlled:
- Resource parameters
- Instance selection
For the public apps, resource parameters are usually adjustable through the Memory per job and CPU per job parameters. Setting the values for these will cause the scheduling component to allocate the most optimal instance type given these values. The following is an example from the task page:
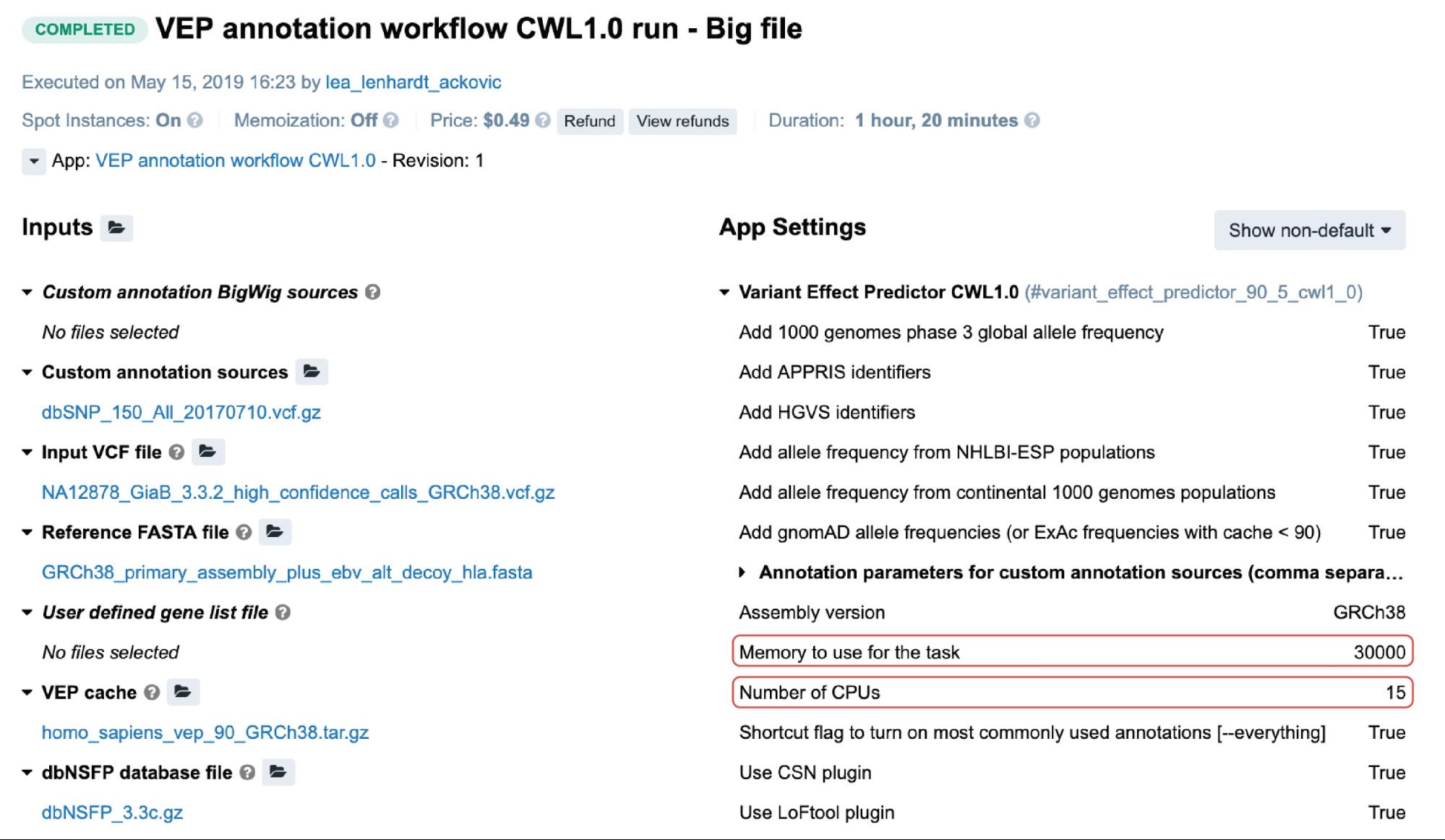
Figure 3. An example of a task containing the two resource parameters. The value for the memory is in megabytes.
In this particular case, c5.4xlarge instance (32.0 GiB, 16 vCPUs) was automatically allocated based on the two highlighted resource parameters (Figure 3). If we take a moment to examine the other instance types which may fit this requirement, we may note that c4.4xlarge comes with 30.0 GiB and 16 vCPUs, which are the values closer to the given ones. However, the platform’s scheduling component also takes into account the price, and since c5.4xlarge is slightly cheaper than c4.4xlarge it was given precedence.
Another way to control the computational resources is to explicitly choose an instance type from the Execution Settings panel on the task page:
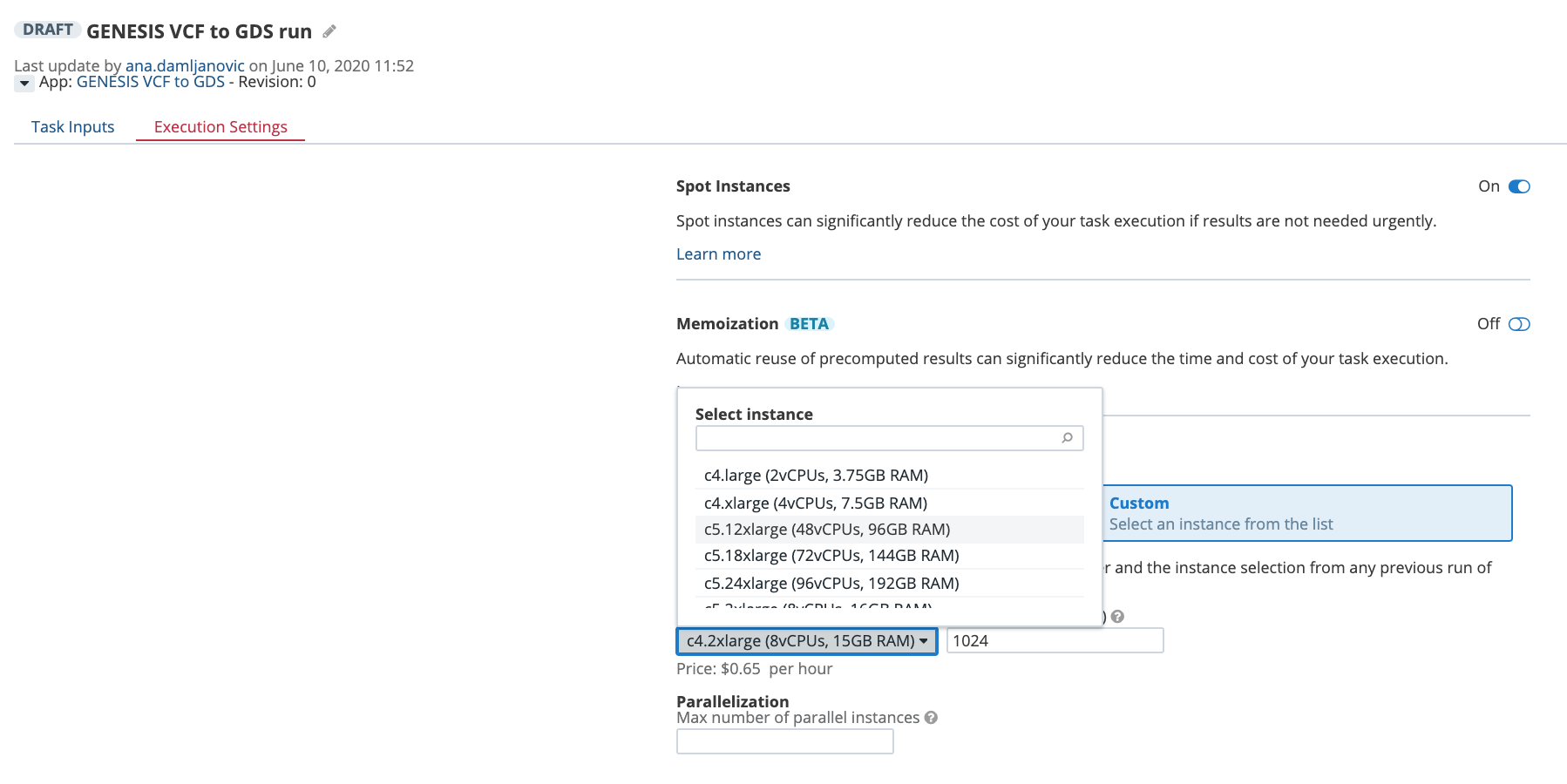
Figure 4. Selecting an instance type from the drop-down menu on the task page.
Learn about Instance Profiles
To be able to get the most out of your computational resources, it is essential to know how your tools behave in different scenarios, like when using different input parameters or different data file sizes. One of the most efficient ways to catch these patterns and use them to optimize future analyses is to monitor resources profiles.
The following is an example of the figures accessible from the Instance metrics panel found within stats & logs page (View stats & logs button is located in the upper right corner on the task page):
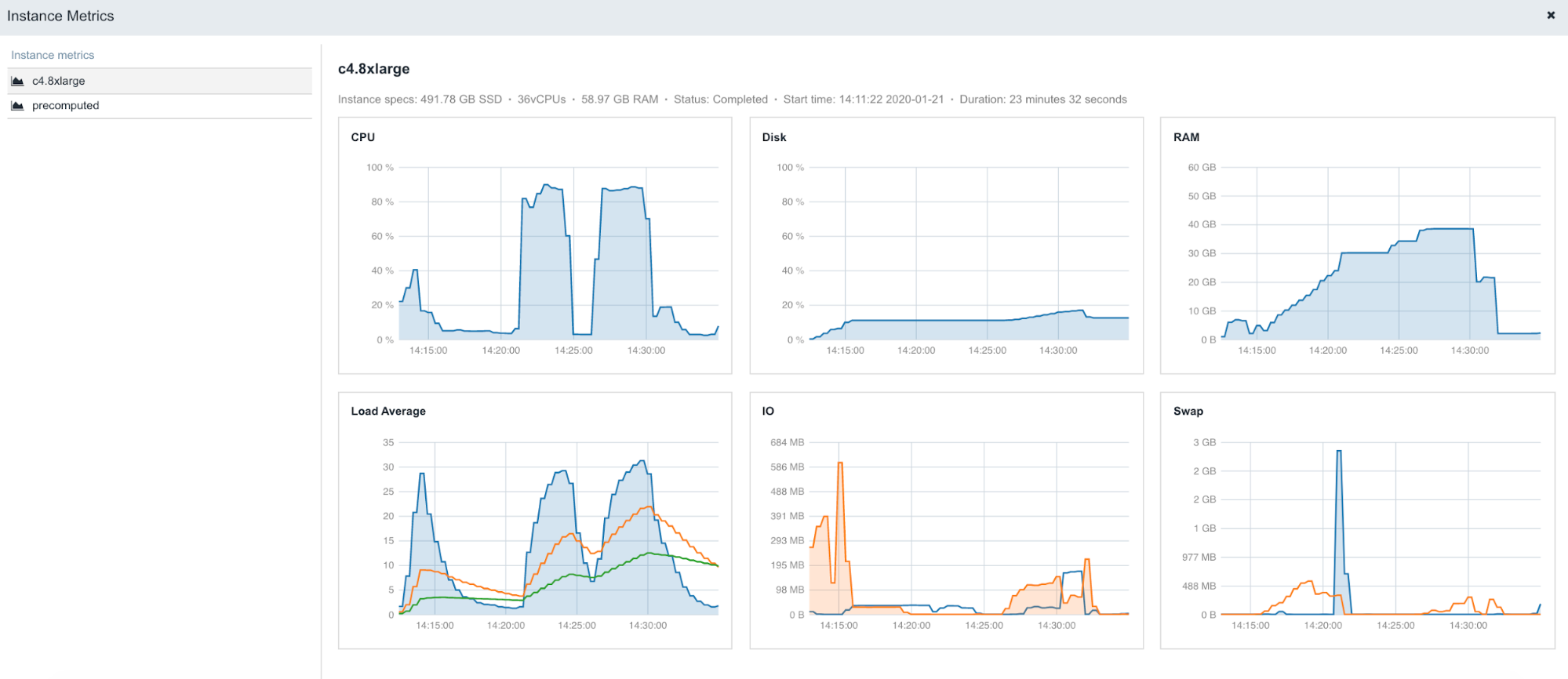
Figure 5. An example of instance profiles when running STAR 2.5.4b on Amazon c4.8xlarge (36 CPUs, 60GB of memory).
To learn more about Instance metrics, visit the related documentation page in our Knowledge Center.
Scale up with Batch Analysis
Batch analysis is an important step when analyzing larger datasets such as TOPMed studies. We use batch analysis to create separate, analytically-identical tasks for each given item (either a file or a group of files depending on the batching method).
Two batching modes are available:
- Batch by File
- Batch by File metadata
In the Batch by File scenario, a task will be created for each file selected in the batched input. In the Batch by File Metadata scenario, a task will be created for each group of files that correspond to a specified value of the metadata (Figure 6).
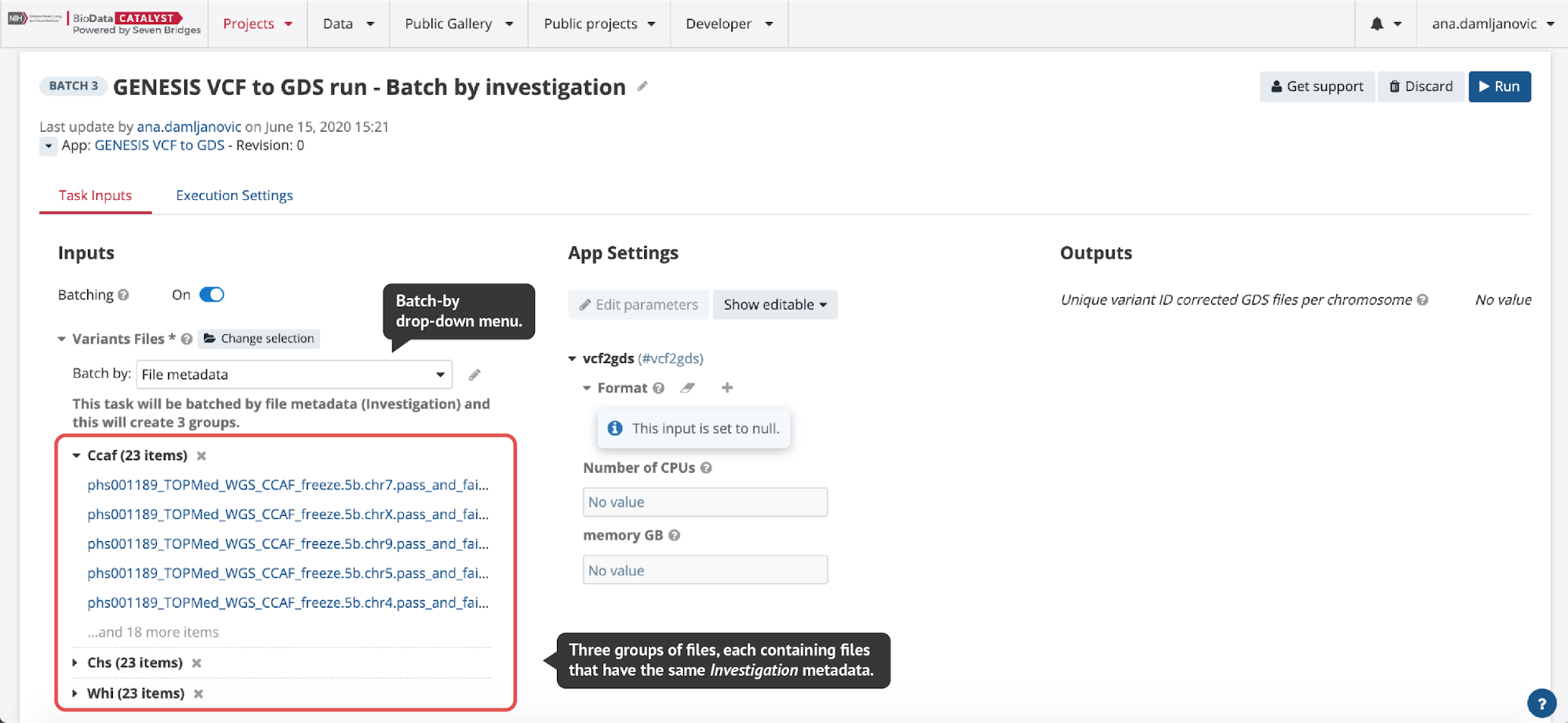
Figure 6. When File metadata is selected from the drop-down menu on the task page, a pop-up window is opened where users can select the metadata they want to batch by. Here, variant calls in VCF format from three different TOPMed studies are converted to GDS format and by selecting Investigation metadata, three tasks are created automatically for each study (investigation).
Batch analysis is quite robust to errors or failures, in that failures in one task will not affect other tasks within the same batch of analysis. In other words, if we process multiple samples using batch analysis, each sample will have its own task. If one task fails, the other tasks will continue to process and can complete successfully, and we will still be able to complete the analysis of most samples. However, we can alternatively decide to process multiple samples using another feature called Scatter (learn more on the following page), and have all the samples processed within a single task. In contrast to batch analysis, a failure of any part of the scatter task (i.e. a job for an individual sample), will cause the entire task to fail and thus the analysis for all samples will fail. In summary, batch analysis highly simplifies the process of creating a large number of the same tasks while allowing independent, simultaneous executions.
Important: Before starting large scale executions with Batch Analysis, please refer to the “Computational Limits” section below.
Parallelize with Scatter
Many common analyses will benefit from highly parallel executions, such as analyzing files from multiple TOPMed studies. The first factor that will determine the parallelization strategy is whether you want to execute the same tool multiple times within one computational instance, or use multiple, independent instances to run the whole analysis in parallel for different input files (i.e. batch analysis). The feature that enables the former is called Scatter.
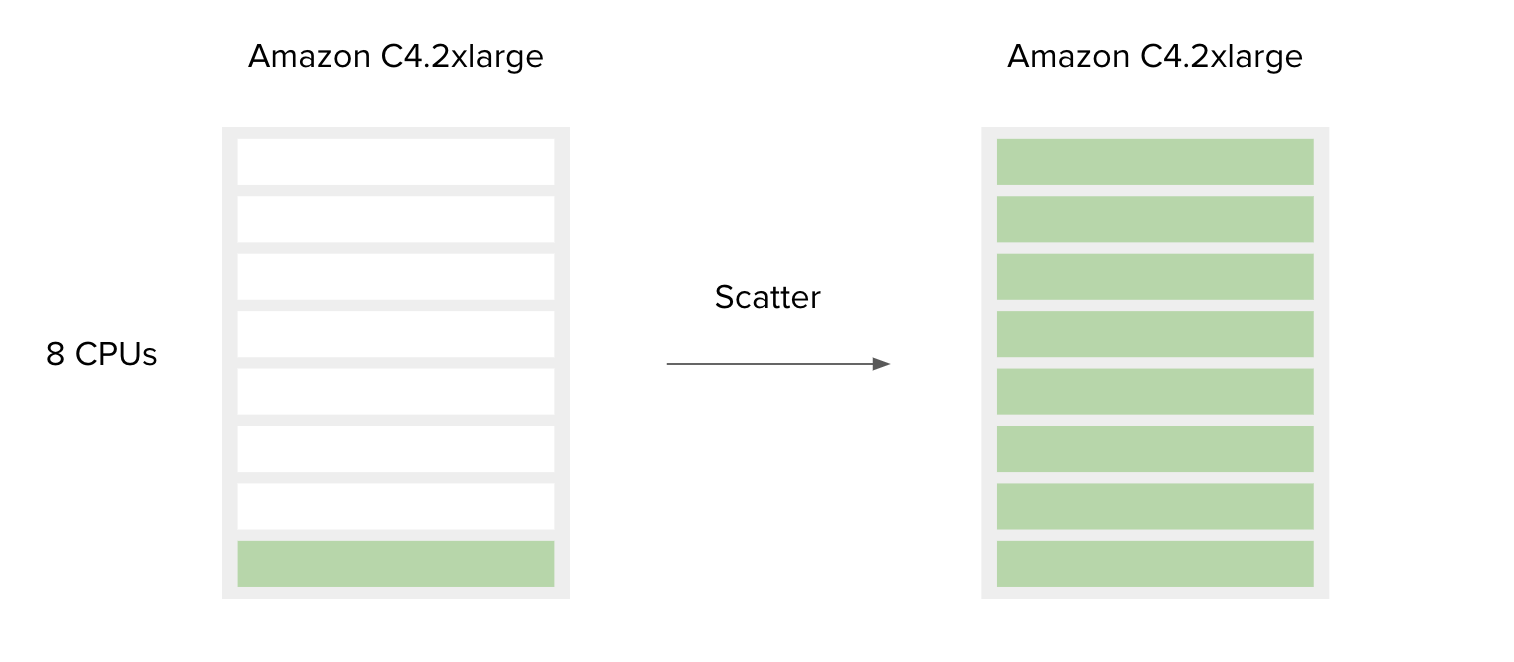
Figure 7. Simplified schematic explaining how scatter affects resource utilization. Green bars represent CPUs in use. If we run only one job that requires one CPU, the other CPUs will remain unused (left), whereas if we run 8 such jobs, all CPUs will be utilized (right). The cost in both scenarios will be the same.
The following is a simple example to accompany Figure 7.
SBG Decompressor is a simple app used for extracting files from compressed bundles. It requires only one CPU and 1000 MB of memory to run. By default, the Amazon c4.2xlarge instance is chosen by the platform since the requirements for CPU and memory of this tool are less than or equal to the c4.2xlarge capacities. This means that if you have only one file to decompress, most of the resources on the c4.2xlarge instance will remain unused. However, if you have multiple files you would like to process, the best way to do this is to use the Scatter feature, and by doing so utilize the additional instance resources. By design, scattering will create a separate job (green bar) for each file (or a set of files, depending on the input type) provided on the scattered input.
Another common application of the scatter feature is seen when using the HaplotypeCaller tool to call SNPs and indels across large genomic intervals (e.g. entire chromosome set). Instead of searching through the whole genome, the HaplotypeCaller tool has an option which allows for defining specific genomics regions (e.g. intervals) within which variants will be called. With this in mind, it is possible to set up the processing such that each chromosome is run independently from each other. This is a great example of a task that will benefit from scattering, as you can run the tool in parallel across all chromosomes. By choosing the “include_intervals” input from the Scatter box (Figure 8), the tool will perform scattering by the genomic intervals input, which will result in an independent job for each provided interval file (usually BED or TXT format which contains interval name along with the corresponding start and end genomic coordinates).
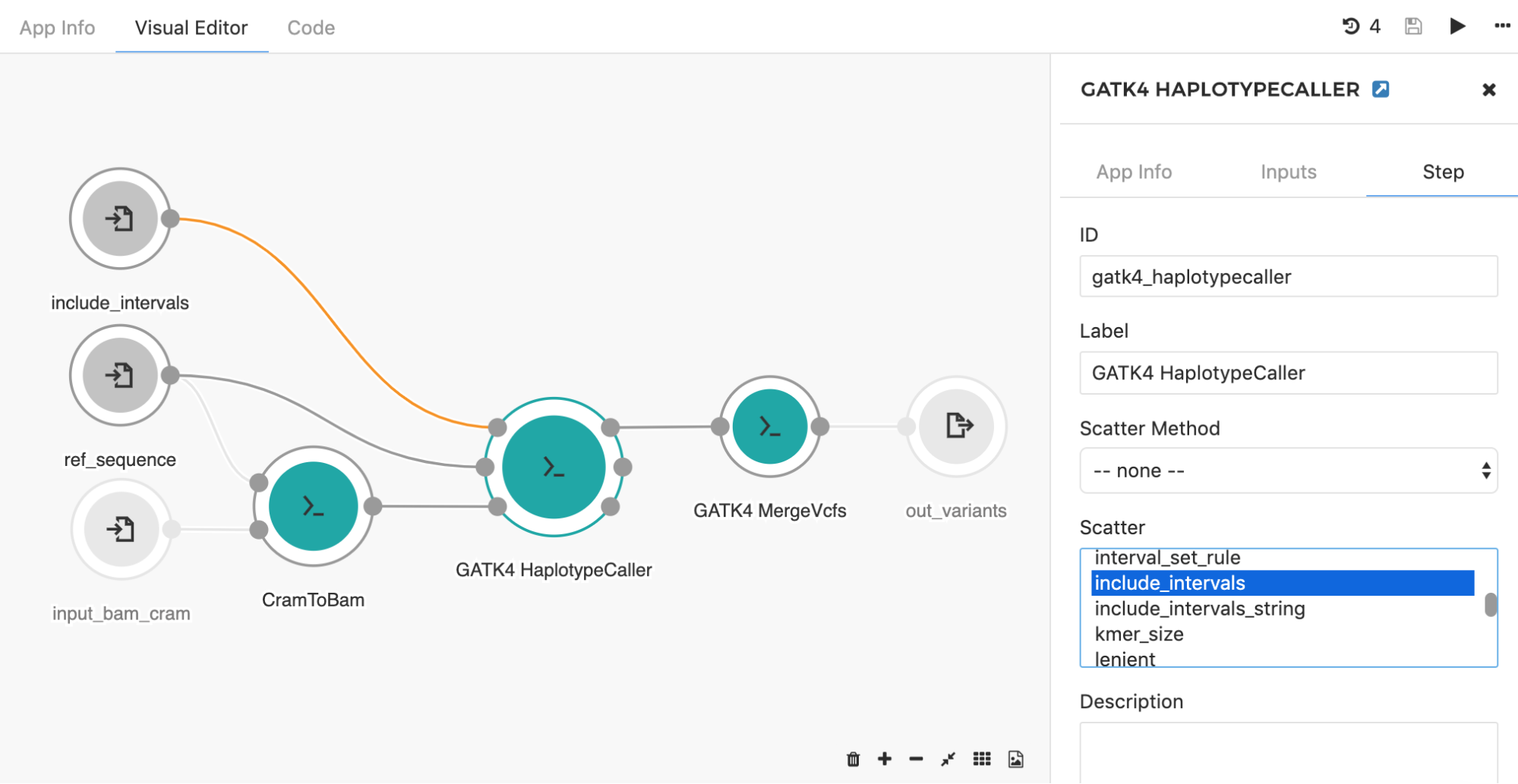
Figure 8. To set scattered input, double click on the tool of interest and select Step in the right-hand panel. The input you want to scatter can be selected under the “Scatter” section. Additionally, if that particular input is connected directly to the input node (there are no preceding tools in between), the input node needs to be modified (also by double clicking on the node) to accept an array of objects originally defined (e.g. array of files instead of one file). Once the input node is adjusted, the connection will turn orange. Here, GATK4 HaplotypeCaller input include_intervals is scattered, meaning that the independent job will be executed for each file provided on this input.
There are situations where we want to create one job per multiple input files. To accomplish this, we need a way to bin the input files into a series of grouped items. A good example of this is when an input sample has paired-end reads, and we want to create jobs for each sample, not each read. In this case, SBG Pair by Metadata tool (available as a public app) can be used to create the group for each pair, and this tool should be inserted upstream of the tool on which you will apply scatter in your workflow.
If you recall the comparison between Batch Analysis and Scatter from the previous section, it may appear that these features are alternatives to one another. Even though they are intended for somewhat similar purposes, Scatter and Batch Analysis are mostly used as complementary features – they can be applied to the same analysis, like for a HaplotypeCaller example above. The workflow already contains the HaplotypeCaller tool which is scattered across intervals. If we run the workflow and batch by input_bam_cram input, we create an individual task for each BAM/CRAM input file.
Another example of Batch Analysis and Scatter working together is the above-mentioned run in which we convert VCF files to GDS format (Figure 6). The GENESIS VCF to GDS workflow is already scattered, meaning that multiple conversions for each input file are performed in a single task. By also using Batch Analysis, we can speed up the conversion to support running a large number of input files and ensure enough disk space on the instance for each batch.
Configuring default computational resources
In a previous section, we reviewed controlling the computational resources from the task page, where you can choose an instance for the overall analysis on-the-fly. This may be exactly what you need in many cases, however, customizing the workflow for a specific scenario or optimizing it with Scatter will probably require setting up default instance types. This can be configured from the tool/workflow editor by using the instance hint feature. This feature allows you to 1) select the appropriate resources for the individual steps (i.e. tools) in a workflow, and 2) define the default instance for the entire analysis (either a single tool or a workflow).
The creator of the analysis can choose to set the default instance if they are confident that their analysis will work well on a particular instance and they don’t want to rely on the user settings or the platform scheduler. However, if the default instance is not set, the user can select the instance for the analysis from the task page (as described above), or the user can leave it to the platform scheduler to pick the right instance based on the resource requirements for the individual tool(s).
If you are looking to set up an instance hint (i.e. choose a specific instance type) on the particular tool in the workflow, you can do so by entering the editing mode, then double-clicking on the node and selecting Set Hints button under the Step tab as follows:
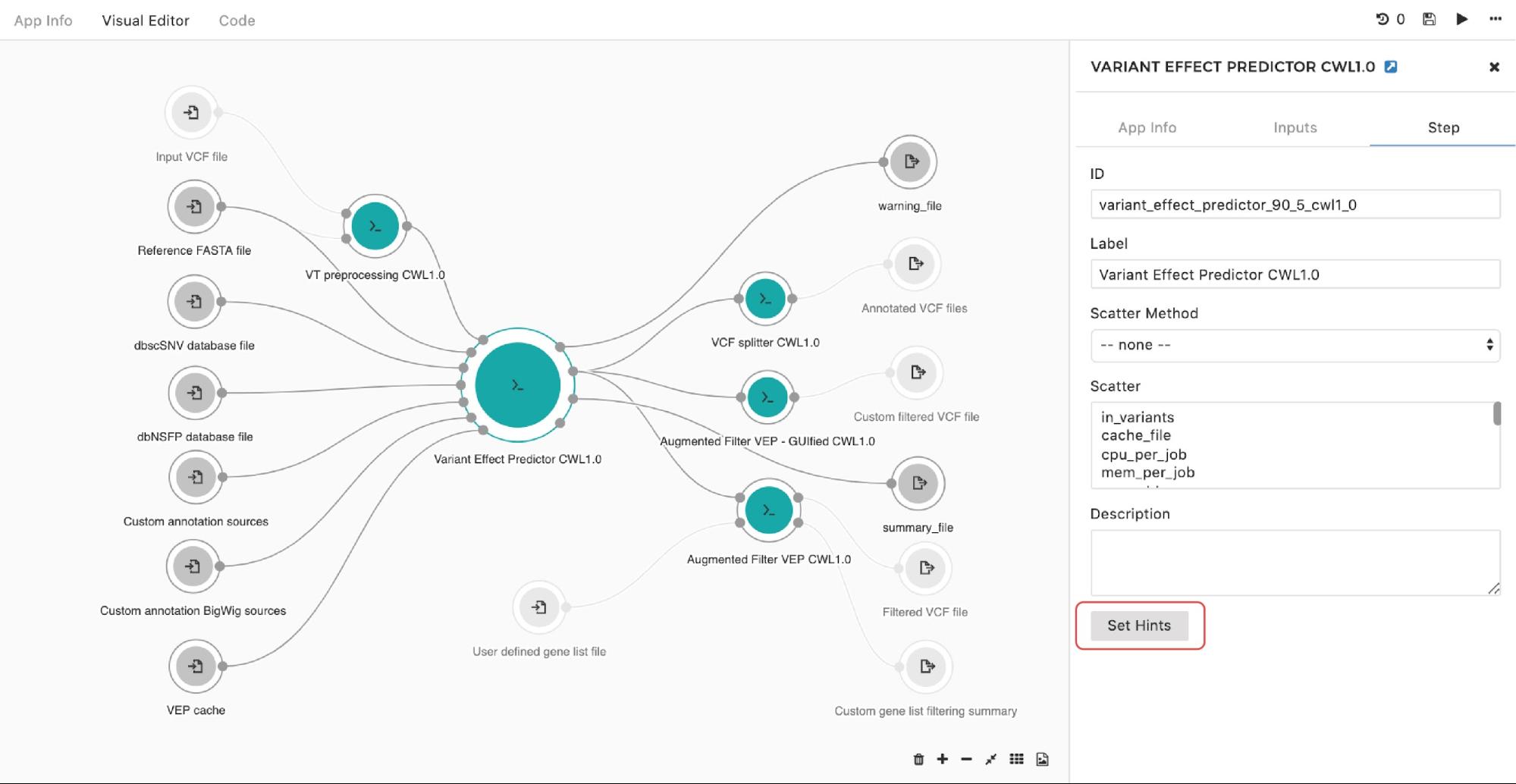
Figure 9. Setting instance hints for a particular app in the workflow.
A configuration pop-up window will appear and you can choose instance types. The same goes for managing the instance hints for the whole workflow. The only difference is that we get to the pop-up window through the drop-down menu in the upper right corner in the editor as shown in Figure 10:
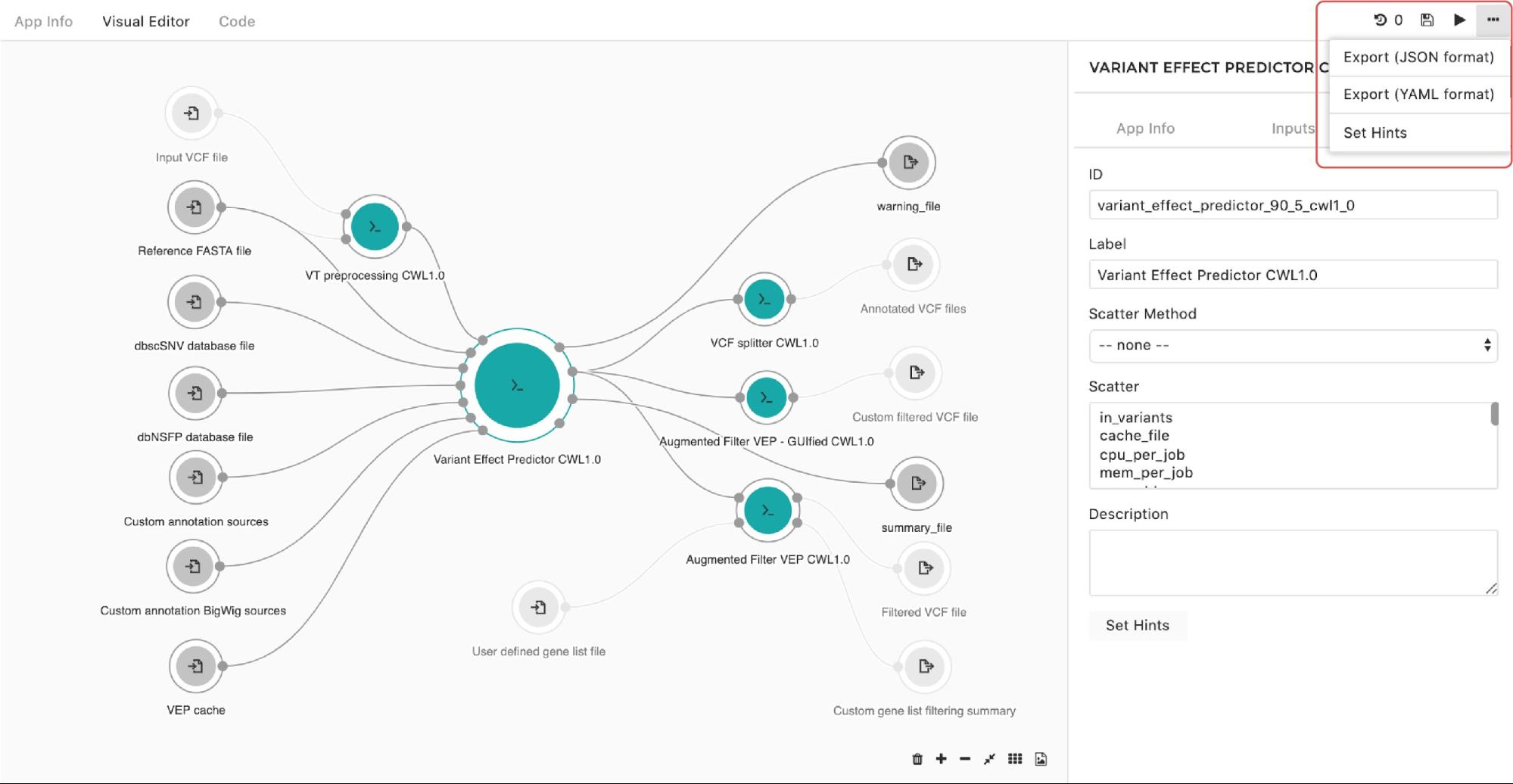
Figure 10. Setting instance hints for the whole workflow.
Finally, to set the instance hints for an individual tool, edit the tool and scroll down to the HINTS section shown in Figure 11:
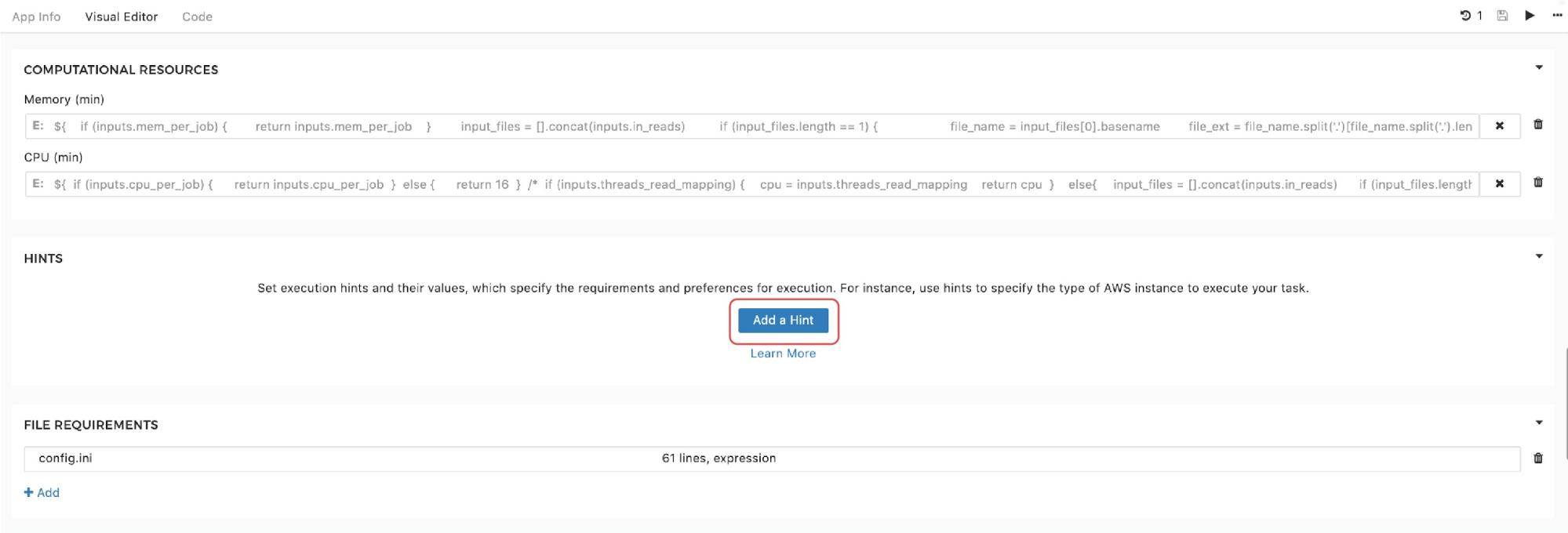
Figure 11. Setting instance hints within tools.
If you recall the resource parameters description, you may wonder what happens if the resource parameters are set and an instance hint is configured within the same tool. In this case, the scheduler will prioritize the instance hint value and will allocate the instance based on that information. However, the information about resource parameters needed for one job is not completely ignored. Namely, if we are about to run the workflow, and the set-up is such that multiple jobs can be executed in parallel on the same instance (i.e. we use Scatter feature), resource parameters are also taken into consideration. They are not used for the instance allocation, but they determine the number of different jobs that can be “packed” together for simultaneous runs on the given instance. We provided a detailed explanation of the scatter feature that enables this in a previous section.
Further analysis and interpretation of your Results
On BioData Catalyst powered by Seven Bridges, you can further analyze your data by leveraging Python notebooks or R interactive capabilities by using the Data Studio feature. With the Data Studio, you can easily access the files in your project for use in an R- or Python-based environment, without the need to download them to your local machine, and with the added flexibility of choosing computational resources.
Getting started
To access the Data Studio:
- In a project on the Platform navigate to the Data Studio tab.
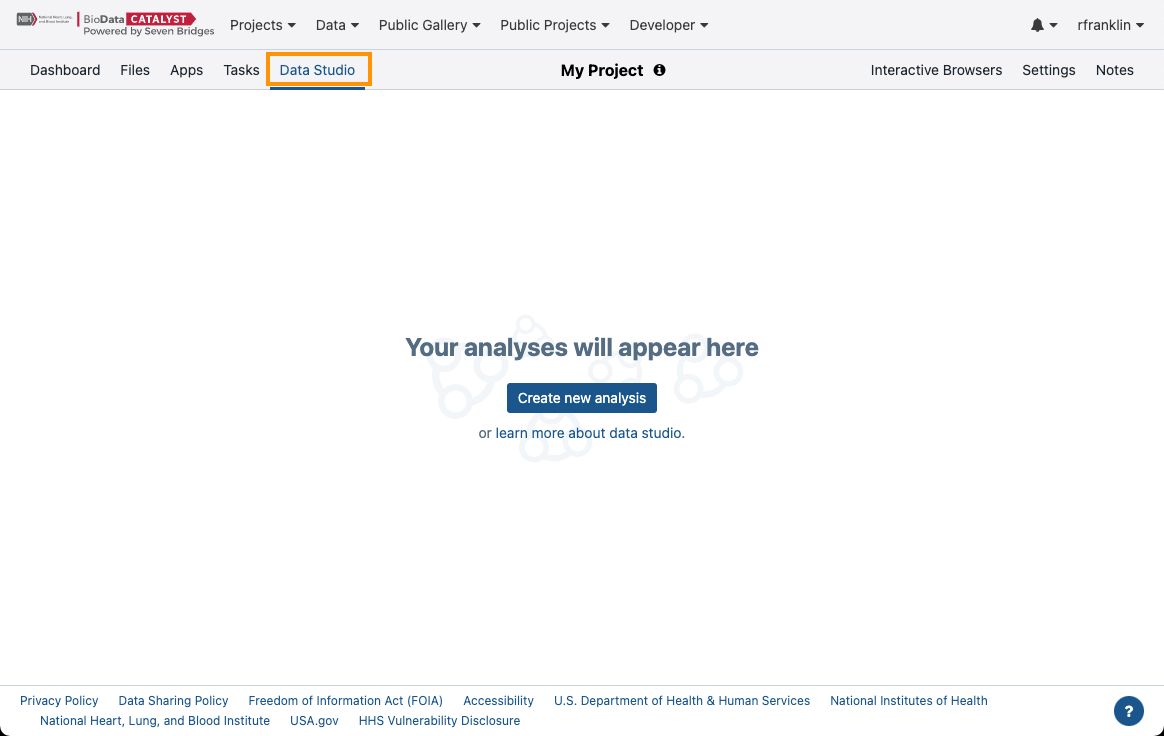
Figure 12. Navigating to the Data Studio.
- Click Create new analysis, which will start an analysis set-up dialogue box (Figure 13)
There are three computing environments available (Figure 13):
- JupyterLab
- RStudio
- SAS Studio
There are several Amazon Web Services (AWS) or Google instance types that you can choose from. If you selected an AWS location at the time of project creation, then you will see AWS instance options, likewise for when Google is set as the project location. You can choose one of two Docker images with pre-installed Python and R packages. The SB Data Science - Python 3.6, R 3.4 image contains the most common packages needed for scientific analysis and visualizations. In addition, you can configure Suspend time as a safety mechanism to enable automatic termination of the instance after a period with no activity. You can turn off this feature completely if you plan a longer interaction and you do not want to risk termination. We recommend that you carefully check the Location and Suspend Time settings before you start the analysis, especially if you are running an existing one which already has its own configuration (Figure 14).
After you set up the preferences, you can click on the Start button to spin up the instance. It may take several minutes for your instance to initialize.
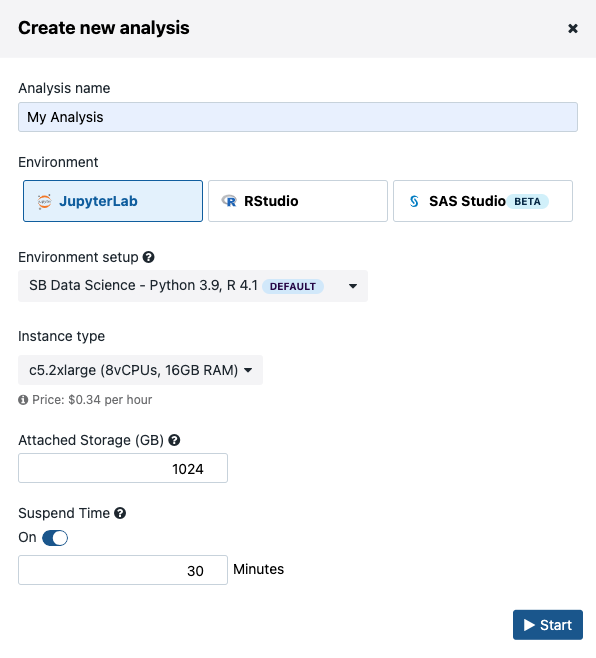
Figure 13. Setting up the interactive analysis in Data Studio.
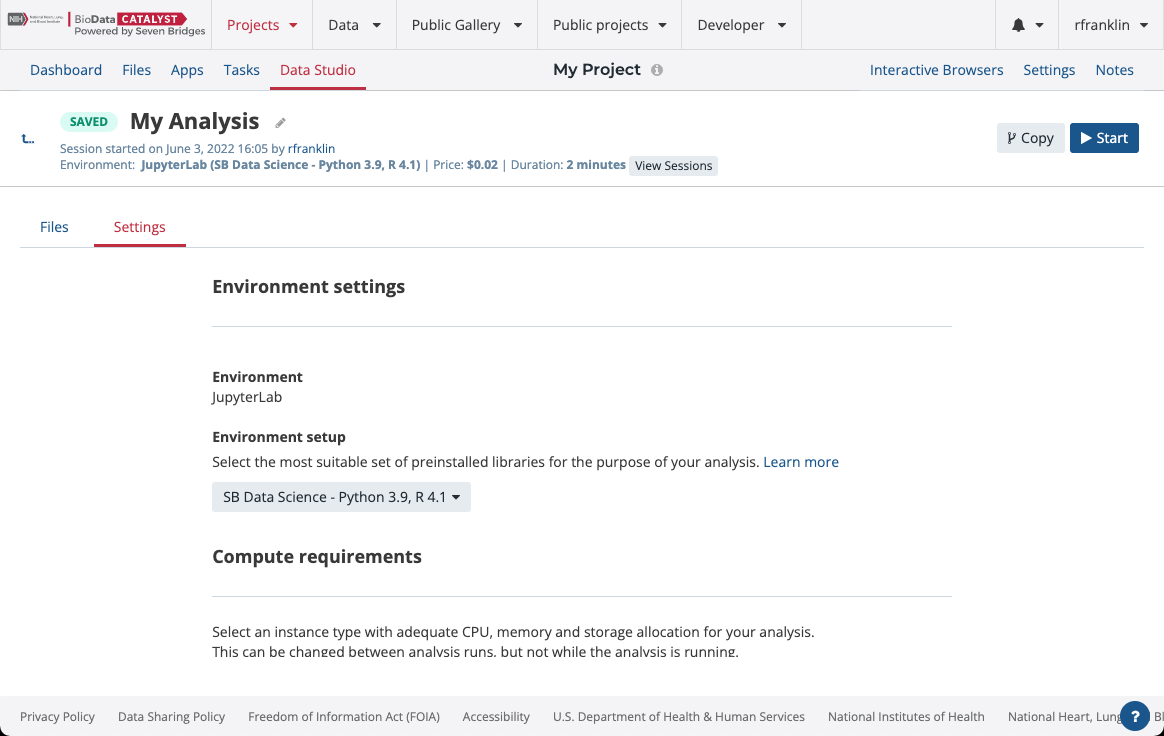
Figure 14. Analysis settings panel. The computational requirements can be edited when the analysis is inactive.
JupyterLab environment
Once the instance is ready, the editor will open. From there, you can move forward with your analysis (Figure 16):
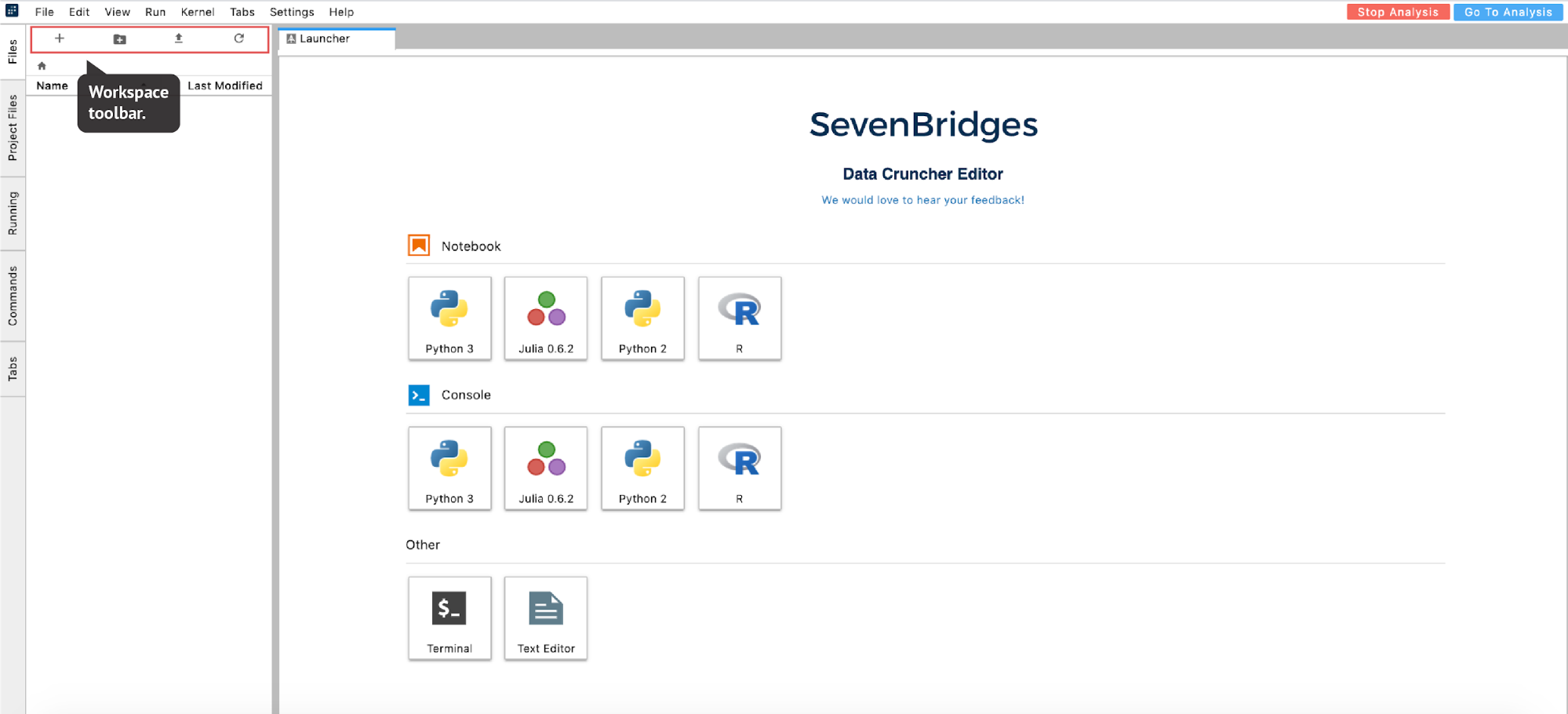
Figure 16. JupyterLab landing page. The list of analysis files on the left is still empty.
Accessing the files
The files you can analyze within the notebook are:
- files present in the analysis – either files uploaded directly in the analysis workspace (your home folder for your interactive analysis) or files produced by the interactive analysis itself, and
- files present in the project
The list of files available in the analysis is displayed in the left-hand panel under the Files tab. This is a list of items in the /sbgenomics/workspace directory, which is the default directory for any work that you do during your session. To control the content in this directory (create new folder, upload files etc.), you can use the workspace toolbar located above this panel (Figure 16).
However, you are likely interested in using an interactive analysis to access the data from your project. There are two ways to get the path from a file found within your project:
- From GUI – click on the file you want to analyze in the Project Files tab (this action copies the path to clipboard) and paste the path in the notebook (Figure 17)
- List all the files in the
/sbgenomics/project-filesdirectory and choose the one you are interested in (see cell [4] in Figure 17).
This path is immutable across different projects. Therefore, if you copy the analysis to another project and you have referenced your file this way, it will work the same way in the new project.
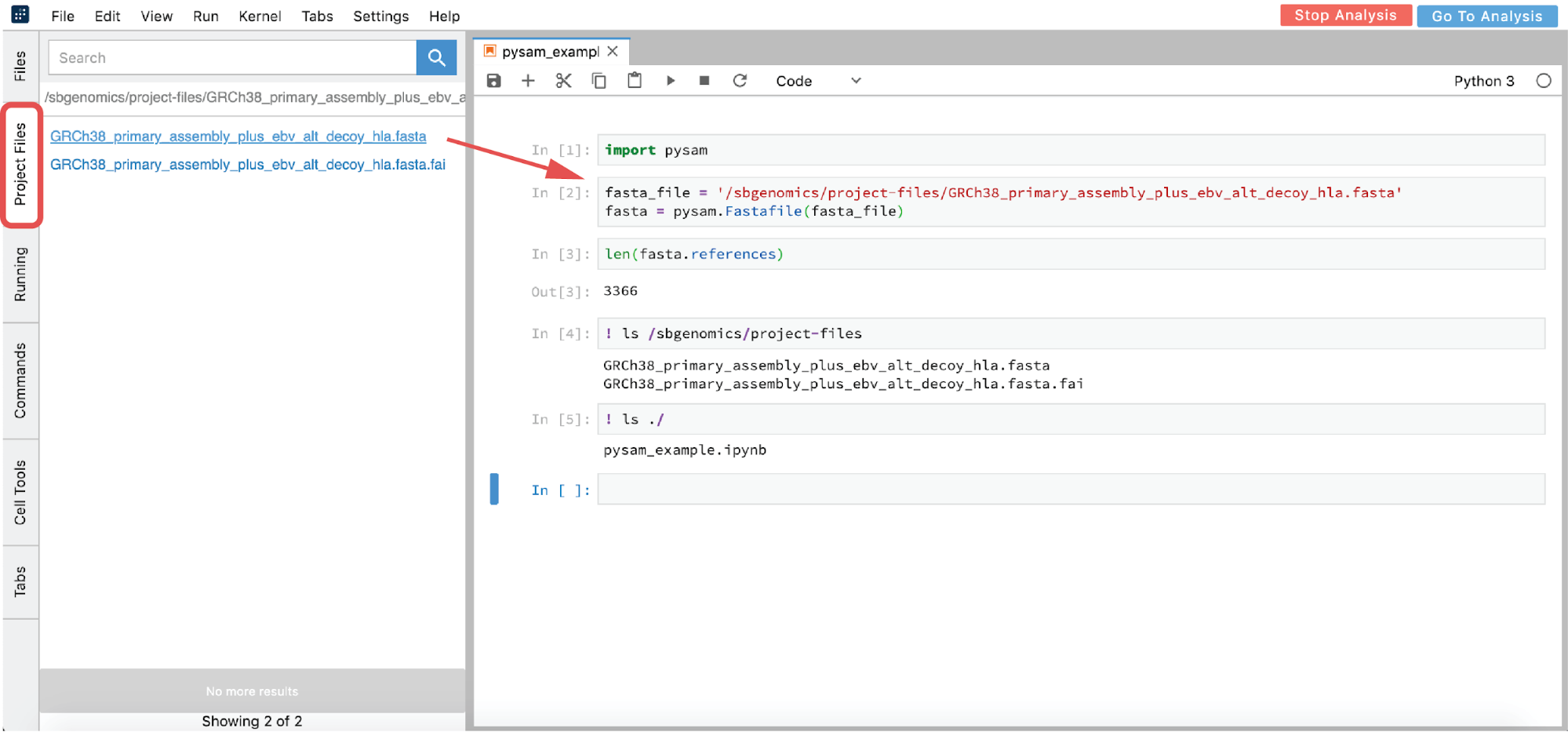
Figure 17. Python 3 Notebook example.
As you may have noticed from the previous example, we used “!” in two notebook cells to switch from Python to shell interpreter and hence denote that these cells should be executed as shell commands. If there is a need to use shell more intensively, like for installation purposes or similar, the notebook environment can become impractical. Fortunately, there is an option to mitigate this: you can open up a terminal from the launcher page (Figure 16).
Saving the created files
Finally, when you are done with the analysis and you want to save the results to your project, go to the Files tab, right-click on the file(s) and select Save To Project. Alternatively, the files can be saved to the project by copying them to /sbgenomics/output-files directory from the terminal. Please note that only smaller files (e.g. .ipynb files) will continue to live in the analysis after it has stopped. Hence, all other needed files need to be saved before you decide to terminate the session.
RStudio environment
In case you prefer to conduct your analysis using RStudio, you should follow the identical procedure from above (Figures 13 and 15) with an exception of selecting RStudio in the dialogue box shown in Figure 13.
Accessing and saving the files in RStudio
Unlike the JupyterLab environment, RStudio environment doesn’t offer convenient file handling from the GUI. However, the same paths that we mentioned in previous sections are used in RStudio:
- Project files directory:
/sbgenomics/project-files - Workspace directory:
/sbgenomics/workspace - Output directory:
/sbgenomics/output-files
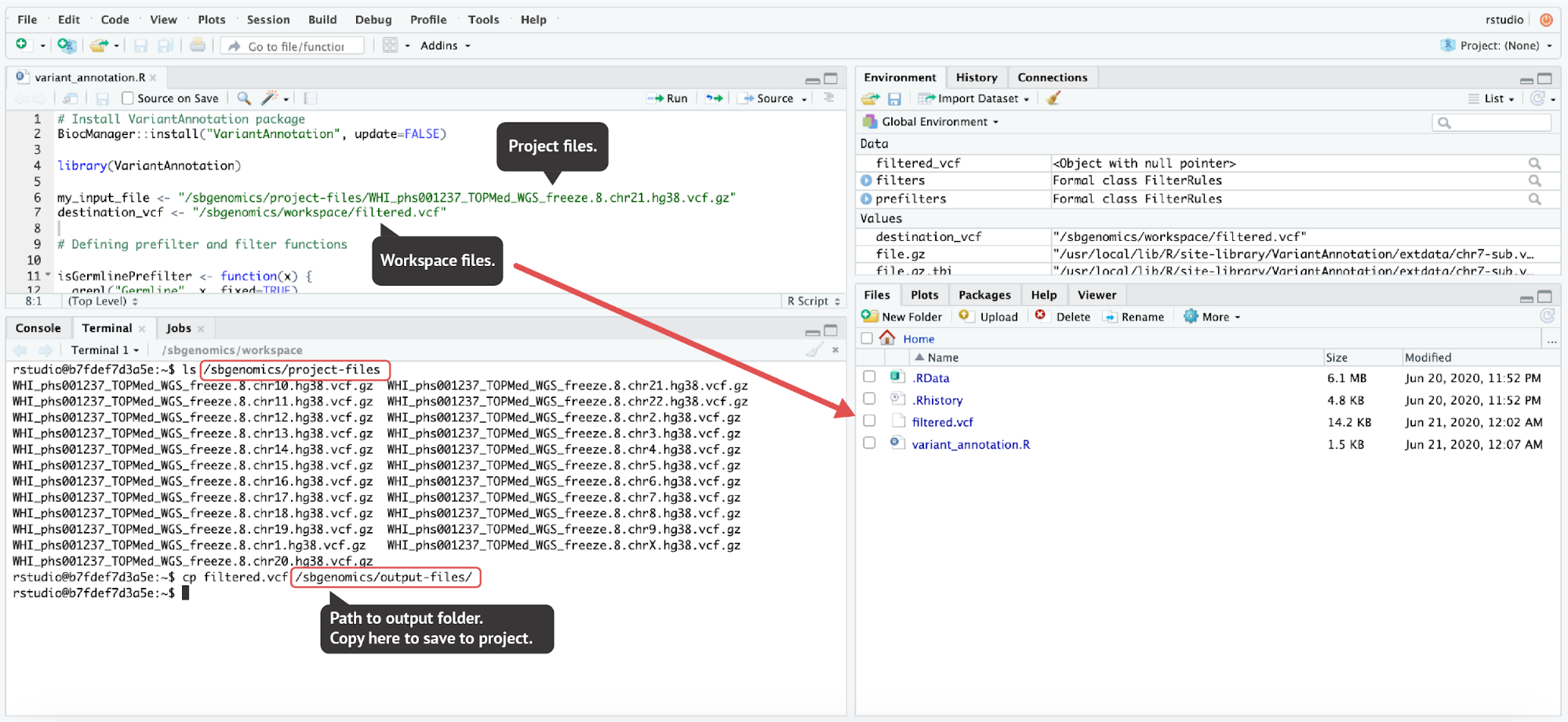
Figure 18. To demonstrate the file handling in RStudio, we performed a simple VCF filtering provided in the VariantAnnotation package vignette. The resulting filtered VCF file was then saved to the project by copying the filtered.vcf file to the output directory (the last command executed in the terminal). The code used here can be found in the Appendix.

Further Reading
Data Studio documentation:
About Data Studio]
YouTube demo:
https://www.youtube.com/watch?v=LP53HNyG9gs&t=6m35s
Computational Limits
As you prepare to run a large number of simultaneous tasks, it is good to be aware of the Platform limitations.
Seven Bridges limits the maximum number of tasks that a user can run simultaneously due to limits for the cloud providers on the total number of instances that can be allocated at the same time on the platform. This limitation helps ensure that there are enough computational resources for all platform users. Users can currently run a maximum of 80 parallel tasks. Therefore, if you create a project and trigger a batch analysis containing 100 tasks, 80 tasks will run immediately and the remaining 20 tasks will be in “QUEUED” status until some of the 80 tasks finish.
If you need to run at a higher throughput, please reach out to the Seven Bridges Support team, so we can work together to find the best solution.
Storage Pricing
Users are not charged for files that are part of the hosted datasets on BioData Catalyst. Therefore, when you copy the files from the Data Browser to your project, you will not be charged for storing those files. The files from these hosted datasets are located in a cloud bucket maintained by NHLBI. These files are referenced in your project, while the physical file remains in the NHLBI cloud bucket.
A similar logic applies to all files: if you copy a file from one project to another, that file is only referenced in the second project, without creating a physical copy and without additional storage expenses. If you create multiple copies of a file and then delete the original file, the copies will still remain and the file will still accrue storage costs until ALL copies are deleted.
In addition to computation, storage costs are made up of the cost for storing derived files or any files that you have uploaded. It is important to note that this applies as long as you keep the files in your projects. To minimize storage costs, it is recommended to check on your files after each project phase is completed and discard any files which are not needed for the future analysis. Note that intermediate files can always be recreated by running the analysis again, and that the cost to re-create these files can sometimes be less than storing the files in your project. As noted in the previous paragraph, multiple copies of a file are charged only once. Consequently, when deleting files which have a copy in other projects, you will still accrue storage expenses until all the copies are removed.
If you have questions about moving your files to manage storage costs, please contact us at [email protected] to learn more about streamlining the process. For more information about storage and computational pricing, visit our Knowledge Center.
Appendix
Using the API
There are numerous use cases in which you might want to have more control over your analysis flow, or simply want to automate custom, repetitive tasks. Even though there are a lot of features enabled through the platform GUI, certain types of actions will benefit from using the API.
We will provide a couple of API Python code snippets for the purpose of this manual. These code snippets have proven to be useful for mitigating most frequent obstacles when running large scale analyses on the Seven Bridges platforms.
Before you proceed, note that all examples are written in Python 3.
Install and configure
To be able to run Seven Bridges API, you should first install sevenbridges-python package:
pip install sevenbridges-python
After that, you can play around with some basic commands, just to get a sense of how the Seven Bridges API works. To be able to initialize the sevenbridges library, you’ll need to generate your authentication token and plug it into the following code:
import sevenbridges as sbg
api = sbg.Api(url='https://api.sb.biodatacatalyst.nhlbi.nih.gov/v2', token='<TOKEN_HERE>')
Managing tasks
Here is a simple code for fetching all task IDs from a given project and printing the status metrics:
import collections
# First you need to provide project slug from its URL:
test_project = 'username/project_name'
# Query all the tasks in the project
# Query commands may be time sensitive if there is a huge number of expected items
# See explanation below
tasks = api.tasks.query(project=test_project).all()
statuses = [t.status for t in list(tasks)]
# counter method counts the number of different elements in the list
counter = collections.Counter(statuses)
for key in counter:
print(key, counter[key])
The previous code should result in something like this:
ABORTED 1
COMPLETED 8
FAILED 1
DRAFT 1
It is of great importance to stop here for a moment and provide an explanation of API rate limits.
The maximum number of API requests, i.e. commands performed on the api object, is 1000 requests in 5 minutes (300 seconds). An example of this command is the following line from the previous code snippet:
tasks = api.tasks.query(project=test_project).all()
By default, 50 tasks will be fetched in one request. However, the maximum number is 100 tasks and it can be configured as follows:
tasks = api.tasks.query(project=test_project, limit=100).all()
In other words, you would have to wait for the renewed requests for a couple of minutes if the project contained more than 100,000 tasks. This scenario almost never happens in practice since it is unusual to have this number of tasks in one project. However, this number can be easily reached for the project files to which the same rule applies. The next few code examples illustrate handling files.
Managing files
Let’s see how we can perform some simple actions on the project files. The following code will print out the number of files in the project:
files = api.files.query(project=test_project, limit=100).all()
len(list(files))
If we now want to read the metadata of a specific file we can run the following:
files = api.files.query(project=test_project, limit=100).all()
for f in list(files):
if f.name == 'phg001287.v1.TOPMed_WGS_WHI_v2.genotype-calls-vcf.WGS_markerset_grc38.c2.HMB-IRB-NPU.tar.gz':
my_meta = f.metadata
for key in my_meta:
print(key,':', my_meta[key])
The output will look like this:
Consent : HMB-IRB-NPU
DbGap accession : phs001237
Study accession :
Study accession with consent :
Study with consent : phs001237.c2
Freeze : freeze5
investigation : WHI
As mentioned earlier, the majority of datasets hosted on the platform have immutable metadata. However, if you have your own files and want to add or modify metadata, you can do so by using the following template:
files = api.files.query(project=test_project, limit=100).all()
for f in list(files):
if f.name == 'example.bam':
f.metadata['foo'] = 'bar'
f.save()
my_meta = f.metadata
for key in my_meta:
print(key,':', my_metadata[key])
Bulk operation: reducing the number of requests
As you can see, f.save() is called each time we want to update the file with new information. This means that the new API request is generated and if we recall that there is a limit of 1,000 requests in 5 minutes, we realize that we can easily hit this number. To solve this issue we can use bulk methods:
all_files = api.files.query(project=test_project, limit=100).all()
all_fastqs = [f for f in all_files if f.name.endswith('.fastq')]
changed_files = []
for f in all_fastqs:
if f is not None:
f.metadata['foo'] = 'bar'
changed_files.append(f)
if changed_files:
changed_files_chunks = [changed_files[i:i + 100] for i in range(0, len(changed_files), 100)]
for cf in changed_files_chunks:
api.files.bulk_update(cf)
In this example, we chose to set metadata only for the FASTQ files found within the project. We perform that by triggering only one API request per 100 files – we first split the list of files into chunks of at most 100 files and then run api.files.bulk_update() method on each chunk.
To learn about other useful bulk operations, check out this section in the API Python documentation.
Managing batch tasks
Another point that might appear necessary is batch analysis handling. If there had been batch tasks in the project which we used for printing out task status information in one of the previous examples, we would not have gotten metrics which include children tasks statuses as well. To be able to query children tasks, we need a couple of additions to our API code.
First, we see how we can check which tasks are batch tasks:
test_project = 'username/project_name'
tasks = api.tasks.query(project=test_project).all()
for task in tasks:
if task.batch:
print(task.name)
What usually happens is that we want to automatically rerun the failed tasks within batch analysis. Here is how we can filter out failed tasks for the given batch analysis and rerun those tasks with an updated app:
test_project = 'username/project_name'
my_batch_id = 'BATCH_ID'
my_batch_task = api.tasks.get(my_batch_id)
batch_failed = list(api.tasks.query(project=test_project,
parent=my_batch_task.id,
status='FAILED',
limit=100).all())
print('Number of failed tasks: ', len(batch_failed))
for task in batch_failed:
old_task = api.tasks.get(task.id)
api.tasks.create(name='RERUN - ' + old_task.name,
project=old_task.project,
# App example: user-name/demo-project/samtools-depth/8
# You can copy this string from the app's URL
app=api.apps.get('username/project_name/app_name/revision_number'),
inputs=old_task.inputs,
run=True)
print('Running: RERUN - ' + old_task.name)
As you can see, there is no need to specify inputs for each task separately – we can automatically pass those inputs from the old tasks. However, this only relates to re-running the tasks in the same project. If you want to re-run the tasks in a different project, you will need to write a couple of additional lines to copy the files and apps.
Collecting specific outputs
Finally, when you are happy with your analysis and want to fetch and further examine specific outputs, you can refer to the next example. Here we share the code for copying all the files belonging to a particular output node from all successful runs to another project. The code can be easily modified to fit your purpose, e.g. renaming the files, deleting them if you want to offload the storage, providing those as inputs to another app, etc. The code may appear a bit more complex because it contains error handling and logging, but the iteration through the tasks and collecting the files of interest is fairly simple.
The resulting code will look like this:
import sevenbridges as sbg
import time
from sevenbridges.http.error_handlers import (rate_limit_sleeper, maintenance_sleeper, general_error_sleeper)
import logging
logging.basicConfig(level=logging.INFO)
time_start = time.time()
my_token = '<INSERT_TOKEN>'
# Copy project slug from the project URL
my_project = 'username/project_name'
# Get output ID from the "Outputs" tab within "Ports" section on the app page
my_output_id = 'output_ID'
api = sbg.Api('https://api.sb.biodatacatalyst.nhlbi.nih.gov/v2', token=my_token, advance_access=True,
error_handlers=[rate_limit_sleeper, maintenance_sleeper, general_error_sleeper])
tasks_queried = list(api.tasks.query(project=my_project, limit=100).all())
task_files_list = []
print('Tasks fetched: ', time.time() - time_start)
for task in tasks_queried:
if task.batch:
ts = time.time()
children = list(api.tasks.query(project=my_project,
parent=task.id,
status='COMPLETED',
limit=100).all())
print('Query children tasks', time.time() - ts)
for t in children:
task_files_list.append(t.outputs[my_output_id])
elif task.status == 'COMPLETED':
task_files_list.append(task.outputs[my_output_id])
print('Outputs from all tasks collected: ', time.time() - time_start, '\n')
fts = time.time()
for f in task_files_list:
print('Copying: ', f.name)
# Uncomment the following line if everything works as expected after inserting your values
# f.copy(project='username/destination_project')
print('\nAll files copied :', time.time() - fts)
print('All finished: ', time.time() - time_start)
There are two major cases – either we run into the batch task in which case we need to iterate through all completed children tasks, or we simply run into an individual task and only need to check if the status is “COMPLETED”. If any of these two conditions are satisfied, we collect the output of interest (lines 19 to 30) and then loop through those to copy the files (lines 35 to 38).
If you have any questions or need help with using API, feel free to contact our support.
RStudio VariantAnnotation Example
In case you would like to test the RStudio environment on Data Studio, you can quickly run the flowing code. Alternatively, you can adjust the code to take one of the VCFs from the TOPMed studies as input. In that case, you will need to create a VCF tabix index in advance (by using the Tabix Index app) and plug its path into the code below.
# Install VariantAnnotation package
BiocManager::install("VariantAnnotation", update=FALSE)
library(VariantAnnotation)
destination_vcf <- "/sbgenomics/workspace/filtered.vcf"
# Defining prefilter and filter functions
isGermlinePrefilter <- function(x) {
grepl("Germline", x, fixed=TRUE)
}
notInDbsnpPrefilter <- function(x) {
!(grepl("dbsnp", x, fixed=TRUE))
}
## We will use isSNV() to filter only SNVs
allelicDepth <- function(x) {
## ratio of AD of the "alternate allele" for the tumor sample
## OR "reference allele" for normal samples to total reads for
## the sample should be greater than some threshold (say 0.1,
## that is: at least 10% of the sample should have the allele
## of interest)
ad <- geno(x)$AD
tumorPct <- ad[,1,2,drop=FALSE] / rowSums(ad[,1,,drop=FALSE])
normPct <- ad[,2,1, drop=FALSE] / rowSums(ad[,2,,drop=FALSE])
test <- (tumorPct > 0.1) | (normPct > 0.1)
as.vector(!is.na(test) & test)
}
prefilters <- FilterRules(list(germline=isGermlinePrefilter, dbsnp=notInDbsnpPrefilter))
filters <- FilterRules(list(isSNV=isSNV, AD=allelicDepth))
file.gz <- system.file("extdata", "chr7-sub.vcf.gz", package="VariantAnnotation")
file.gz.tbi <- system.file("extdata", "chr7-sub.vcf.gz.tbi", package="VariantAnnotation")
# Uncomment the following two lines and insert paths to your files
# file.gz <- "/sbgenomics/project-files/WHI_phs001237_TOPMed_WGS_freeze.8.chr21.hg38.vcf.gz"
# file.gz.tbi <- "/sbgenomics/project-files/WHI_phs001237_TOPMed_WGS_freeze.8.chr21.hg38.vcf.gz.tbi"
tabix.file <- TabixFile(file.gz, yieldSize=10000)
filterVcf(tabix.file, "hg19", destination_vcf, prefilters=prefilters, filters=filters, verbose=TRUE)
filtered_vcf <- readVcf(destination_vcf, "hg19")
For more information on the VariantAnnotation package, visit the Bioconductor page here.
Updated 2 months ago