AWS Cloud storage tutorial
On this page:
- Overview
- Procedure
- Prerequisites
- Step 1: Register an S3 bucket as a volume
- 1a: Create an IAM (Identity and Access Management) user
- 1b: Authorize this IAM user to access your bucket
- 1c: Register a bucket
- Step 2: Make an object from the bucket available on the Platform
- 2a: Launch an import job
- 2b: Check if the import job has completed
- Step 3: Move a file from the Platform to the bucket
- 3a: Upload a file to a project
- 3b: Move a file from your project on the Platform to the bucket
- 3c: Check if the export job has completed
The Volumes API contains two types of calls: one to connect and manage cloud storage, and the other to import and export data to and from a connected cloud account.
Before you can start working with your cloud storage via the Platform, you need to authorize the Platform to access and query objects on that cloud storage on your behalf. This is done by creating a "volume". A volume enables you to treat the cloud repository associated with it as external storage for the Platform.
You can 'import' files from the volume to the Platform to use them as inputs for computation. Similarly, you can write files from the Platform to your cloud storage by 'exporting' them to your volume. Learn more about working with volumes.
The BioData Catalyst powered by Seven Bridges uses Amazon Web Services as a cloud infrastructure provider. This affects the cloud storage you can access and associate with your Platform account.
For instance, you have full read-write access to your data stored in Amazon Web Services' S3 and read-only access to data stored in Google Cloud Storage.
This short tutorial will guide you through setting up a volume. You'll register your Amazon S3 bucket as a volume, make an object from the bucket available on the Platform, then move a file from the Platform to the bucket.
Once a volume is created, you can issue import and export operations to make data appear on the Platform or to move your Platform files to the underlying cloud storage provider.
In this tutorial we assume you want to connect to an Amazon S3 bucket. The procedure will be slightly different for other cloud storage providers, such as a Google Cloud Storage bucket. For more information, please refer to our list of supported cloud storage providers.
To complete this tutorial, you will need:
- An Amazon Web Services (AWS) account
- One or more buckets on this AWS account
- One or more objects (files) in your target bucket
- An authentication token for the Platform. Learn more about getting your authentication token.
##Step 1: Register an S3 bucket as a volume
To set up a volume, you have to first register an AWS S3 bucket as a volume. Volumes mediate access between the Platform and your buckets, which are local units of storage in AWS.
Register an AWS S3 bucket as a volume through the following steps below.
(Optional You can also provide your KMS ID if you opt to use KMS for your encryption.
###1a: Create an IAM (Identity and Access Management) user
Follow AWS documentation for directions on creating an IAM user.
Be sure to keep your credentials somewhere safe. You can also click Download Credentials to obtain them in a file named credentials.csv.
###1b: Authorize this IAM user to access your bucket
- In the list of IAM users, locate the IAM user you created above. Click the username to configure your options.
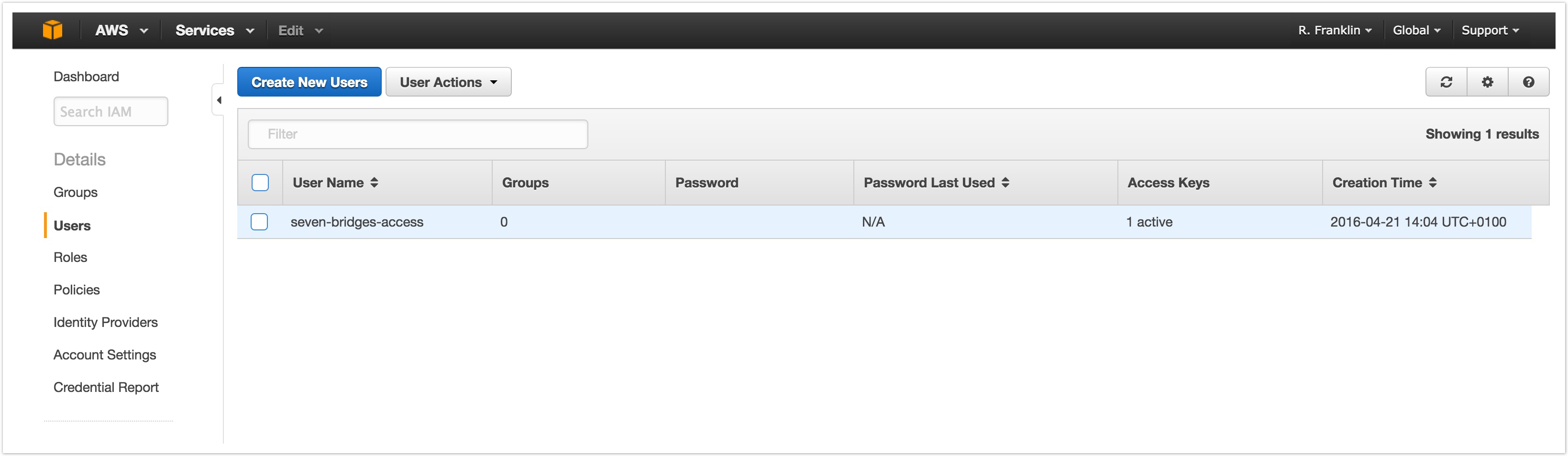
- On the Permissions tab, select Inline Policies then choose click here, as shown below.
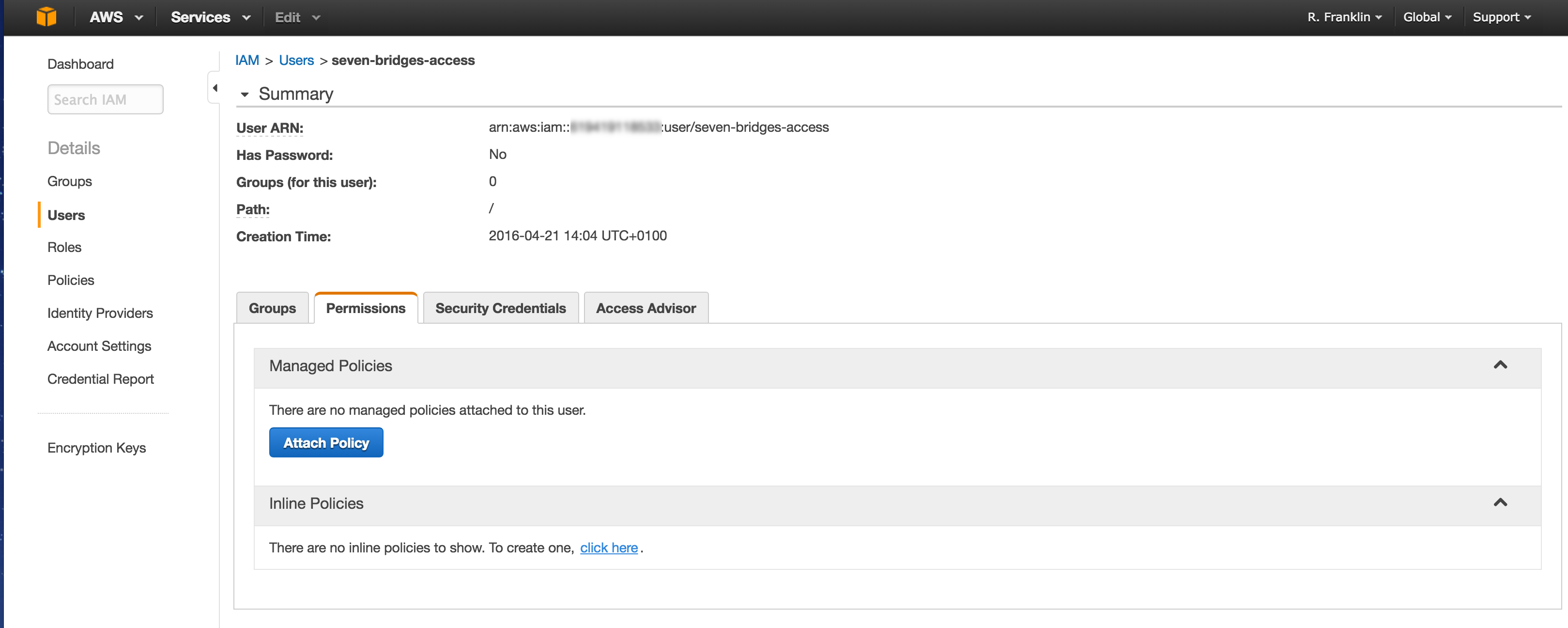
- Choose Custom Policy, then click Select. This will allow you to make a policy specifying the permissions you give to the Platform regarding access to your bucket.
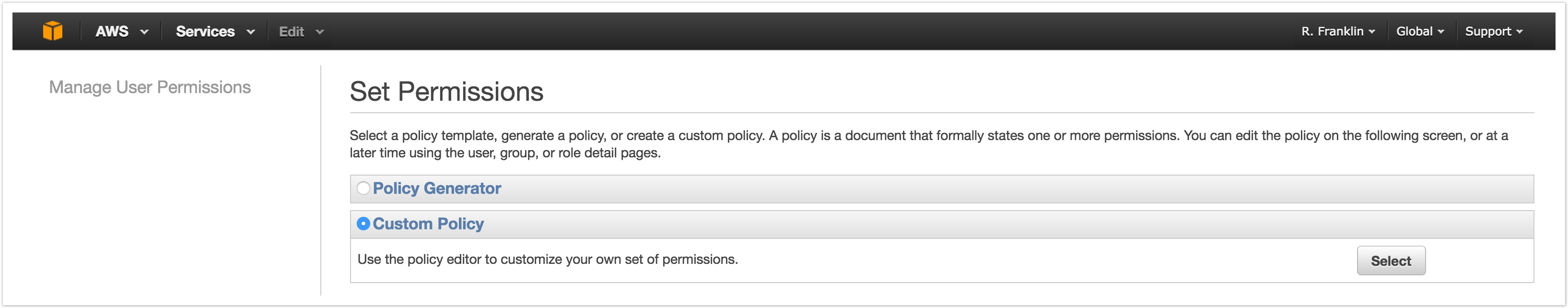
- Enter a descriptive policy name, e.g. sb-access-policy. Note that you can only use alphanumerics and the following characters: +=,.@-_ .
- Then, visit Seven Bridges' AWS policy generator, to generate a custom policy, as shown below.
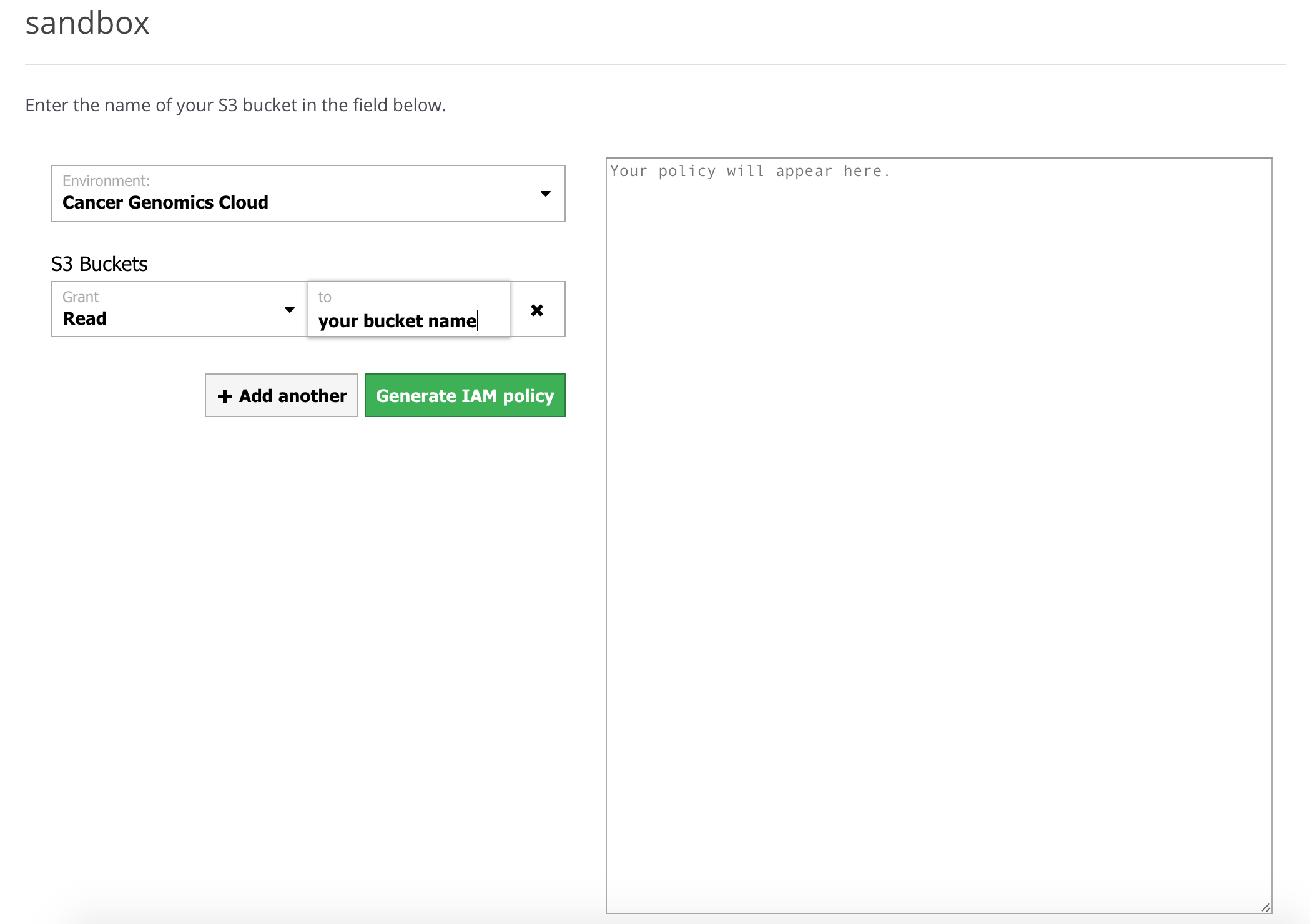
- Select your Cancer Genomics Cloud from the Environment drop-down menu.
- Under Grant, select Read & Write.
- Input your S3 bucket's name under the S3 Buckets section in the box labeled to.
- You can add another S3 bucket by clicking + Add another.
10.Click Generate IAM policy when you are ready. Your policy will appear in the right panel. - Copy and paste it in the policy box in the IAM permissions page on the AWS Console.
- Click Apply policy to save your changes.
###1c: Register a bucket
At this point, you can associate the bucket with your Platform account by registering it as a volume.
To register your bucket as a volume, make the API request to Create a volume, as shown in the HTTP request below. Be sure to paste in your authentication token for the X-SBG-Auth-Token key.
This request also requires a request body. Provide a name for your new volume, an optional description, and an object (service) containing the information in the table below. Specify the access_mode as RW for write permissions or RO for read-only permissions. Be sure to replace BUCKET-NAME with the name of your bucket and substitute in your own credentials.
| Key | Description of value |
|---|---|
typerequired | This must be set to s3. |
bucketrequired | The name of your AWS S3 bucket. |
prefixdefault: empty string | If provided, the value of this parameter will be used to modify any object key before an operation is performed on the bucket. Even though AWS S3 is not truly a folder-based store and allows for almost arbitrarily named keys, the prefix is treated as a folder name. This means that after applying the prefix to the name of the object the resulting key will be normalized to conform to the standard path-based naming schema for files. For example, if you set the prefix for a volume to a10, and import a file with location set to test.fastq from the volume to the Platform, then the object that will be referred to by the newly-created alias will be a10/test.fast". |
endpointdefault: s3.amazonaws.com | The endpoint to use when talking to AWS S3. Note: Volumes associated with buckets hosted on AWS's China (Beijing) region must have endpoint set to s3.cn-north-1.amazonaws.com.cn. Volumes associated with buckets hosted in any other zone may use the default value. |
access_key_idrequired | The access key ID of the IAM user used for operations on this bucket. You will be provided with this when you create the IAM user. |
secret_access_keyrequired | The secret access key of the IAM user to use for operations on this bucket. You will be provided with this when you create the IAM user. |
aws_canned_acl | Specifies an S3 canned ACL to apply when exporting an object to this volume. For more information on the canned ACLs supported by S3, please see the list of canned ACLs in the AWS documentation. |
sse_algorithmdefault: empty | This indicates whether server-side encryption should be enabled. Supported values are:null: do not use server-side encryption;AES256: use Amazon S3-managed keys (SSE-S3).* aws:kms: use Amazon KMSSupport for SSE-C will be added in a later release. For more information on AWS server-side encryption, see the AWS webpage Protecting Data Using Server-Side Encryption. |
POST /v2/storage/volumes HTTP/1.1
Host: api.platform.sb.biodatacatalyst.nhlbi.nih.gov
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
content-type: application/json
{
"name": "tutorial_volume",
"service": {
"type": "s3",
"bucket": "BUCKET-NAME",
"prefix": "",
"credentials": {
"access_key_id": "INSERT ACCESS KEY ID",
"secret_access_key": "INSERT SECRET ACCESS KEY"
},
"properties": {
"sse_algorithm": "AES256"
}
},
"access_mode": "RW"
}
You'll see a response providing the details for your newly created volume, as shown below.
{
"href": "https://api.platform.sb.biodatacatalyst.nhlbi.nih.gov/v2/storage/volumes/rfranklin/tutorial_volume",
"id": "rfranklin/tutorial_volume",
"name": "tutorial_volume",
"access_mode": "RW",
"service": {
"type": "S3",
"bucket": "rfranklin-test-volume",
"prefix": "",
"endpoint": "s3.amazonaws.com",
"credentials": {
"access_key_id": "ACCESS KEY HERE"
},
"properties": {
"sse_algorithm": "AES256"
}
},
"created_on": "2016-06-26T16:44:20Z",
"modified_on": "2016-06-26T16:44:20Z",
"active": true
}
##Step 2: Make an object from the bucket available on the Platform
Now that we have a volume, we can make data objects from the bucket associated with the volume available as "aliases" on the Platform. Aliases point to files stored on your cloud storage bucket and can be copied, executed, and organized like normal files on the Platform. We call this operation "importing". Learn more about working with aliases.
To import a data object from your volume as an alias on the Platform, follow the steps below.
To import a file, make the API request to start an import job as shown below. In the body of the request, include the key-value pairs in the table below.
| Key | Description of value |
|---|---|
volume_idrequired | Volume ID from which to import the file. This consists of your username followed by the volume's name, such as rfranklin/tutorial_volume. |
locationrequired | Volume-specific location pointing to the file to import. This location should be recognizable to the underlying cloud service as a valid key or path to the file. Please note that if this volume was configured with a prefix parameter when it was created, the prefix will be prepended to location before attempting to locate the file on the volume. |
destinationrequired | This object should describe the Platform destination for the imported file. |
projectrequired | The project in which to create the alias. This consists of your username followed by your project's short name, such as rfranklin/my-project. |
name | The name of the alias to create. This name should be unique to the project. If the name is already in use in the project, you should use the overwrite query parameter in this call to force any file with that name to be deleted before the alias is created. If name is omitted, the alias name will default to the last segment of the complete location (including the prefix) on the volume. Segments are considered to be separated with forward slashes ('/'). |
overwrite | Specify as true to overwrite the file if the file with the same name already exists in the destination. |
POST /v2/storage/imports HTTP/1.1
Host: api.platform.sb.biodatacatalyst.nhlbi.nih.gov
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
content-type: application/json
{
"source":{
"volume":"rfranklin/tutorial_volume",
"location":"example_human_Illumina.pe_1.fastq"
},
"destination":{
"project":"rfranklin/my-project",
"name":"my_imported_example_human_Illumina.pe_1.fastq"
},
"overwrite": true
}
The returned response details the status of your import, as shown below.
{
"href": "https://api.platform.sb.biodatacatalyst.nhlbi.nih.gov/v2/storage/imports/5rand0mXYcDQ3xtSHrKuK2jXNDtJhMBN",
"id": "5rand0mXYcDQ3xtSHrKuK2jXNDtJhMBN",
"state": "PENDING",
"overwrite": true,
"source": {
"volume": "rfranklin/tutorial_volume",
"location": "example_human_Illumina.pe_1.fastq"
},
"destination": {
"project": "rfranklin/my-project",
"name": "my_uploaded_example_human_Illumina.pe_1.fastq"
}
}
Locate the id property in the response and copy this value to your clipboard. This id is an identifier for the import job, and we will need it in the following step.
###2b: Check if the import job has completed
To check if the import job has completed, make the API request to get details of an import job, as shown below. Simply append the import job id obtained in the step above to the path.
GET /v2/storage/imports/5rand0mXYcDQ3xtSHrKuK2jXNDtJhMBN HTTP/1.1
Host: api.platform.sb.biodatacatalyst.nhlbi.nih.gov
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
content-type: application/json
The returned response details the state of your import. If the state is COMPLETED, your import has successfully finished. If the state is PENDING, wait a few seconds and repeat this step.
You should now have a freshly-created alias in your project. To verify that a file has been imported, visit this project in your browser and look for a file with the same name as the key of the object in your bucket.
##Step 3: Move a file from the Platform to the bucket
You've successfully created an alias on the Platform for a file in your S3 bucket. You can also move files from the Platform into your connected S3 bucket. This operation is known as 'exporting' to the volume associated with the bucket. Please keep in mind that public files, files belonging to Platform-hosted datasets, archived files, and aliases cannot be exported. For more information, please see working with aliases.
Follow the steps below to move a file from Platform to an object in your bucket.
###3a: Upload a file to a project
Before you can export a file from the Platform, you must upload a file to a project. To upload a file, follow the steps below:
- Upload a file to your project using the command line uploader, the via the visual interface, using an FTP or HTTP(S) server, or the API.
- Locate and copy the file ID. From the various upload mechanisms, you can find the file ID as follows:
- Command line uploader - In the output of the command line uploader, note that the first column in the line that corresponds to the uploaded file. This is the uploaded file's ID.
- Upload via the visual interface - Once the file has uploaded, locate the file in the Files tab of the relevant project. Click on the file's name. A new page with details about your file should open. Locate the last segment of this page's URL, following
/files/. This is the uploaded file's ID. For example, the file ID ofhttps://platform.sb.biodatacatalyst.nhlbi.nih.gov//u/rfranklin/volumes-api-project/files/577d4c35e4b05e75806f2853/is577d4c35e4b05e75806f2853. - FTP or HTTP(S) server - Locate the file's ID in the same way as for files uploaded using the visual interface.
- API - Issue the API request to List all files within a project. The IDs of each file are listed next to the key id in the response body.
###3b: Move a file from your project on the Platform to the bucket
When you export a file from the Platform to your volume, you are writing to your S3 bucket.
Make the API request to start an export job to move a file from the Platform to your bucket, as shown below. In the body of your request, include the key-value pairs from the table below.
| Key | Value |
|---|---|
sourcerequired | This object should describe the source from which the file should be exported. |
filerequired | The Platform-assigned ID of the file for export. |
destinationrequired | This object should describe the destination to which the file will be exported. |
volumerequired | The ID of the volume to which the file will be exported. |
locationrequired | Volume-specific location to which the file will be exported. This location should be recognizable to the underlying cloud service as a valid key or path to a new file. Please note that if this volume has been configured with a prefix parameter, the value of prefix will be prepended to location before attempting to create the file on the volume. |
properties | Service-specific properties of the export. These values override the defaults from the volume. |
sse_algorithmdefault: AES256 | S3 server-side encryption to use when exporting to this bucket. Supported values: AES256 (SSE-S3 encryption)aws:kms* null (no server-side encryption) |
sse_aws_kms_key_id | Provide your AWS KMS ID here if you specify aws:kms as your sse_algorithm. Learn more about AWS KMS. |
POST /v2/storage/exports HTTP/1.1
Host: api.platform.sb.biodatacatalyst.nhlbi.nih.gov
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
content-type: application/json
{
"source": {
"file": "576159f7f5b4e1de6ae9b5f0"
},
"destination": {
"volume": "rfranlin/tutorial_volume",
"location": ""
},
"properties": {
"sse_algorithm": "AES256"
}
}
The returned response details the status of your import, as shown below.
{
"href": "https://api.platform.sb.biodatacatalyst.nhlbi.nih.gov/v2/storage/exports/yrand0mrmxx4Zjr1u781HJaBGOhx02sd",
"id": "yrand0mrmxx4Zjr1u781HJaBGOhx02sd",
"state": "PENDING",
"source": {
"file": "567890abc9b0307bc0414164"
},
"destination": {
"volume": "rfranklin/tutorial_volume",
"location": "output.vcf"
},
"started_on": "2016-06-15T19:17:39Z",
"properties": {
"sse_algorithm": "AES256",
"aws_storage_class": "STANDARD",
"aws_canned_acl": "public-read"
},
"overwrite": false
}
Locate the id property in the response and copy this to your clipboard. This id is the identifier for the export job, and we will use it in the next step to verify that the job has completed.
###3c: Check if the export job has completed
To check the status of your export job, make the API request to get details of an export job. Append the export id you obtained in the step above after the path.
GET /v2/storage/exports/yrand0mrmxx4Zjr1u781HJaBGOhx02sd HTTP/1.1
Host: api.platform.sb.biodatacatalyst.nhlbi.nih.gov
X-SBG-Auth-Token: 3259c50e1ac5426ea8f1273259740f74
content-type: application/json
The returned response details the state of your export. If the state is COMPLETED, your export has successfully finished. If the state is PENDING, wait a few seconds and repeat this step.
Your bucket now contains the file that was uploaded to the Platform in step 1. To verify that a file has been exported, visit your project on the Platform and locate the file you originally uploaded. It should be marked as an alias. This means that the content of the file has been moved from storage on the Platform to your S3 bucket, and that the Platform file record been updated accordingly.
Congratulations! You've now registered an S3 bucket as a volume, imported a file from the volume to the Platform, and exported a file from the Platform to the volume. Learn more about connecting your cloud storage from our Knowledge Center.
Updated over 3 years ago
