Collaborating on the NHLBI BioData Catalyst(R): A Guide for Consortia
Audience
This guide is intended for researchers who are part of a consortium working with the NHLBI BioData Catalyst(R) (BDC) and need a platform solution for data management and sharing across a large number of organizations and individual contributors.
The best practices described here focus on the needs of groups within a consortium (e.g., Data Coordinating Centers, Sequencing Centers, and Analysis Working Groups) to enable large-scale collaboration.
Objective
This guide explains how to use the features and functionality of the BioData Catalyst ecosystem to fulfill the needs of the consortium. In this guide, consortium members and data owners will learn:
- How a consortium can create projects on the BDC-SB platform aligned with the needs of different groups within the consortium;
- How to use the collaboration features to ensure only authorized individuals are part of projects with controlled datasets;
- How to take advantage of the flexibility in cloud location on the platform to align with where individual study centers store their datasets; and,
- How to efficiently manage cloud costs and billing for multiple organizations working together in one consortium.
Overview (The Problem Statement)
As research has become increasingly interdisciplinary, cooperation between various disparate research teams and institutions has become essential. A research consortium is a network of geographically distributed collaborating institutions that aims to perform coordinated scientific research activities. These research consortia are composed of many organizations working together, each with dedicated responsibilities. For example, Study Coordinating Centers (SCCs) generate raw data. The Data Coordinating Centers (DCCs) harmonize datasets from multiple Study Coordinating Centers. Then, the Data Coordinating Center distributes quality controlled, harmonized data. Analysis Working Groups then use these harmonized data sets for research analysis and discovery.
Compared to the individual researcher, operations at a consortium level have their own unique needs. Oftentimes, these consortia manage large datasets, which bring additional challenges to data analysis.
Some example challenges include:
-
Datasets need to be shared between multiple organizations, which often leads to multiple copies of the datasets since individual universities may manage a copy on their local HPCs.
-
Storing and managing large datasets can be cumbersome, requiring resources to manage the data and ensure the latest versions are available, and some universities may lack these resources. Additionally, institutional agreements must be put in place to ensure sensitive controlled datasets are handled appropriately, with only those researchers who have the appropriate credentials being able to access controlled data. For consortia, managing user access to the datasets is typically a manual process.
-
In this vein, members of Analysis Working Groups from different institutions often may not be able to easily share methods and results with each other, which is of critical importance to the efficient flow of data within a consortium.
Within these consortia are numerous groups with unique responsibilities, all working in concert. These groups are involved in data and metadata management, data submission for streamlined analysis, data processing and dissemination, outreach and other activities, and provide the individual building blocks comprising a consortium. An example of the typical research consortium structure is detailed in Figure 1 below.
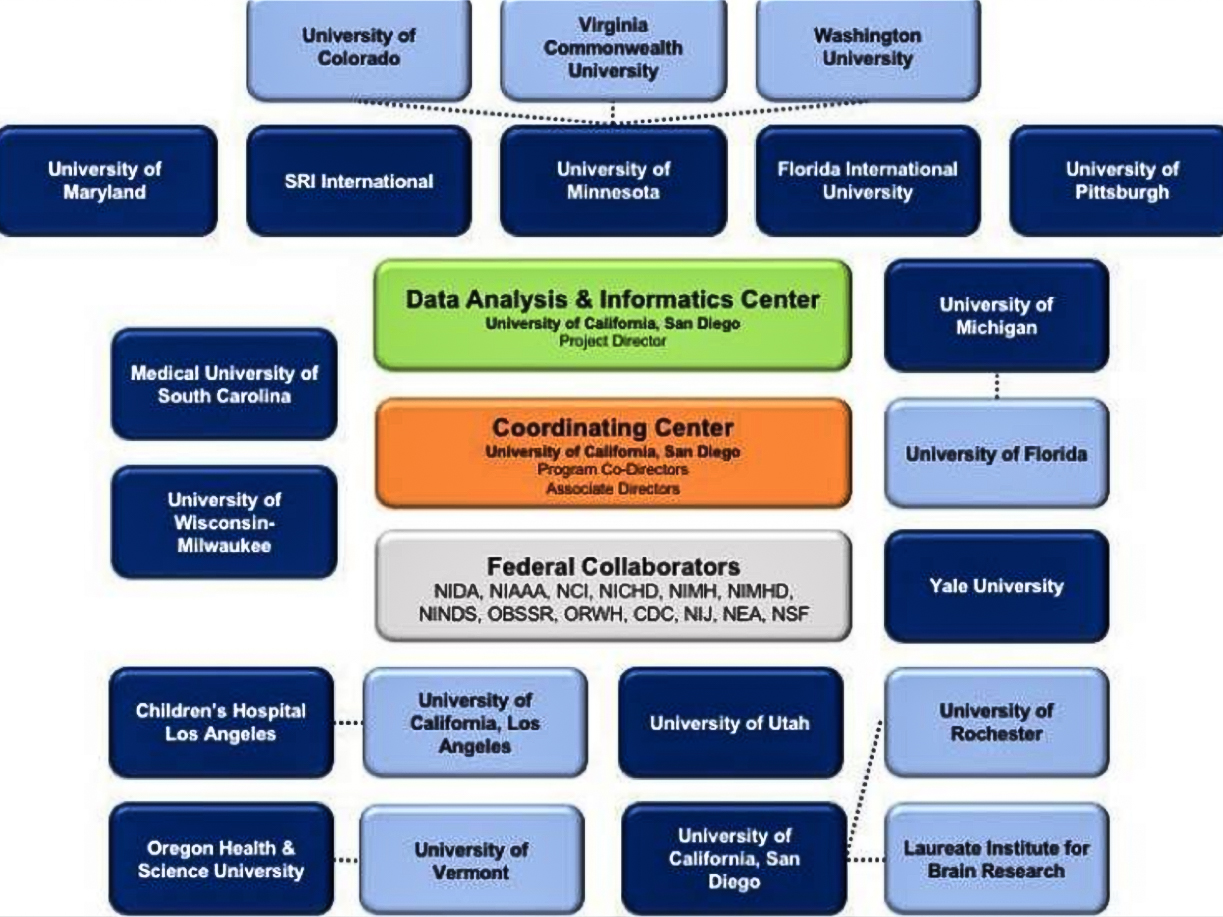
Figure 1: Example organizational diagram of a research consortium. Note the numerous institutions, interrelationships, and various roles within the consortium. Figure adapted from: Auchter, Allison M., et al. "A description of the ABCD organizational structure and communication framework." Developmental Cognitive Neuroscience 32 (2018): 8-15.
Collaborative research on the NHLBI BioData Catalyst(R)
The BioData Catalyst ecosystem is an ideal place for consortia like this to operate. The BioData Catalyst platform provides safe, secure, and efficient data sharing, along with features to manage data access and permissions for numerous collaborators in an organized manner. Additionally, it offers powerful and flexible project options tailored to specific consortia needs.
In the following sections, this guide will discuss each of these main topics in detail, and illustrate how the BioData Catalyst empowers consortia to work more efficiently:
- Projects Map to Purpose: Consortia can organize the work of different groups into particular platform projects.
- Administrative Capabilities for Project Owners: Administrators can set up projects in advance, and then add appropriate members. Membership in the project is limited to those who are approved to access data and those who need to carry out the defined work.
- Cloud Costs and Billing: Cloud costs from consortia projects can be paid for using one or more funding sources, mapping to the grant structure of the consortia.
- Cloud Agnostic: Choose from AWS or Google Cloud to accommodate Study Coordinating Centers that may have their raw data stored on either cloud provider.
Projects map to purpose
With BioData Catalyst, consortia can work together on one platform using multiple distinct workspaces called “projects.” A project is the core building block of the platform and serves as a container for its data, analysis workflows, and results.
Multiple workflow executions can be carried out within a single project. Projects can be used to limit distribution of data and methods to particular individuals based on their role in the consortia. Each project has one or more Project Owners and Project Members. Project Owners are also project administrators.
The project administrator controls which individuals are added as members of the project, as shown in Figure 2 below. The project administrator can also set granular permissions on what the project members can see and do within the project, like whether they can run analyses on the data. In this way, access to the project contents can be restricted to only those individuals who are permitted to access the data.
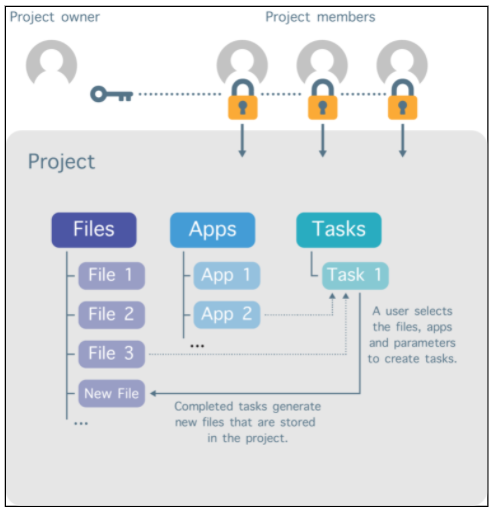
Figure 2: Project Owner and Project Members. The project owner can add collaborators to the project and define permissions.
Consider a hypothetical consortium composed of the following working groups who need to carry out the specified actions:
| Working Group | Members | Action(s) |
|---|---|---|
| Two (2) Study Coordinating Centers | Study PI and a subset of DCC members | Share raw data with the Data Coordinating Center |
| One (1) Data Coordinating Center | DCC members only | Receive raw data from the Study Coordinating Centers QC and harmonize the raw data Share harmonized data back with Study Coordinating Centers Distribute select harmonized data to Analysis Working Groups |
| Two (2) Analysis Working Groups | Analysis Working Groups members only | Receive harmonized data from the Data Coordinating Center Analyze harmonized data and publish results |
Table 1: Breakdown of the hypothetical consortium
Recommendation
We recommend this example consortium use the BioData Catalyst platform to set up several distinct projects that map to purpose. In this way, the various actions of the consortium can be organized within separate projects with only the necessary individuals having access to the project contents.
Figure 3 below shows a conceptual diagram of the types of projects that could be created on the platform to enable the example consortium groups above to work together.
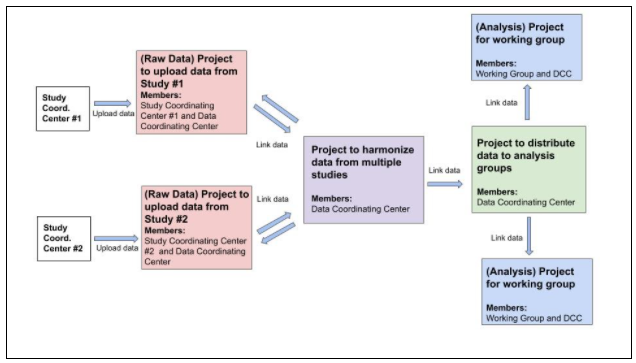
Figure 3: A conceptual diagram for how the consortium can use projects on the BioData Catalyst for data sharing and management.
Seven Bridges recommends the consortia in this scenario set up the following projects:
*The two individual SCCs can upload data to the platform using their own Raw Data Projects (red boxes). Since the DCC are members of the Raw Data Projects as well as the SCC members, this figure as a way of sharing the raw data from each SCC with the DCC. For these groups, administrators will intend on sharing the data with only the corresponding Study PI and a subset of DCC members for each study as required.
-
After data is uploaded to the Raw Data Projects, the DCC members can link the datasets to a separate project: the Harmonization Project (purple box). Notably, this does not create a new copy of the data. The DCC can perform quality control steps and harmonize the data within the Harmonization Project. The DCC can share the harmonized data with the SCCs by linking the harmonized data files back to the Raw Data Projects (indicated by the bidirectional arrows in Figure 3). The SCCs have the option to download the harmonized data back to local computing infrastructure. For this project, the intent is to have all DCC members gain access, which is a separate (but possibly overlapping) group of members from those in the Raw Data Projects. Limiting membership of the Harmonization Project to members of the DCC prevents members of one SCC from accessing data from the other SCC.
-
A Data Distribution Project (green box), once created, will serve as a data repository for the harmonized data from which the DCC members can manage data distribution to the two Analysis Working Groups and their respective members.
-
From the Data Distribution Project, specific files can be added to the two Analysis Projects (blue boxes). Each Analysis Project includes DCC members in addition to those who are tasked with data analysis. The administrators of the Analysis Projects have full control over adding members to the project, thereby allowing them the ability to control who has access to the data for analysis.
In general practice, Seven Bridges recommends that the DCCs map out the platform projects needed for their consortium before having all the individual contributing members set up platform accounts and upload data. A member of the DCC can create all needed projects on the platform and be the owner of all the projects. This gives the DCC full control over who will be added as members of the project with access to the data.
Learn more
Bringing data to the Platform
BioData Catalyst offers a variety of options for the Study Coordinating Center to upload or import data to the platform. For small data uploads, users can drag and drop files from their own computer.
For large data uploads, users can utilize the Command Line Uploader, which can upload at maximum of 250,000 files.
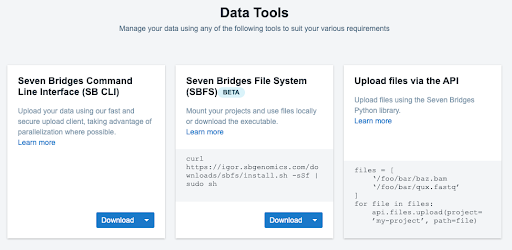
Figure 4: Data Tools on the BioData Catalyst platform
figure4
Additionally, data can be linked from cloud storage buckets on Amazon Web Platform (AWS) and Google Cloud Platform (GCP). If data is stored in a private AWS bucket, users can connect the AWS bucket directly to the Platform using the Volumes feature.
The data will stay in the private bucket and not be physically moved to the BioData Catalyst cloud storage. If data is stored in a private GCP bucket, connect the Google bucket directly to the platform using the Volumes feature.
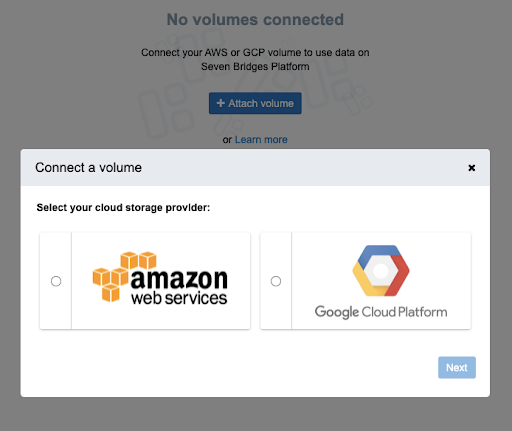
Figure 5: Connecting a volume from your choice of cloud storage provider
Note that data can be linked between projects. This may be of particular interest to members of a DCC, who wish to share files by linking files from one portal to another. Linking these files is achieved through the platform’s visual interface.
Learn more
Making projects findable
The Project tagging feature on the BioData Catalyst platform can help consortia organize and find projects. Project Administrators can assign tags to their projects via the API or through the visual interface. These tags can be used for filtering purposes when browsing all projects, for project categorization, and for general custom organization of projects.
Recommendation
We recommend using these tags in the different consortia projects to enable filtering and better organization.
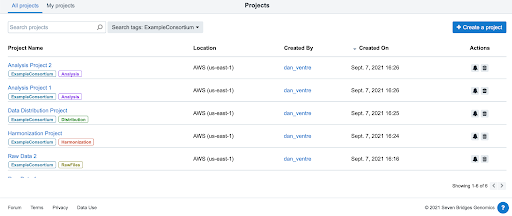
Figure 6: Project Tagging is found by clicking on the “Tags” icon on the Project Dashboard. In this example, eacfigure6h of the projects was tagged with “ExampleConsortium,” allowing for these projects to be filtered quickly on the project dashboard. Additional tags can also be added to each tag as shown above.
Cloud costs and billing
Consortia may have one or more sources of funding (like multiple grants), and need a way to utilize those funding sources on their data management and analysis platform. The BioData Catalyst uses Billing Groups to track cloud costs and facilitate invoicing. Each platform project must have a single billing group defined.
The billing group is set at the time of project creation and can be changed later. Any cloud costs incurred in the project are tracked to the corresponding billing group and then the Seven Bridges team invoices the administrator of the billing group on a monthly basis..
Recommendation
Only admins of a project are allowed to set the billing group and can change it at any time. Only users who are “members” of the billing group are able to select to utilize the billing group for a platform project.
As an example, assume a DCC managed one source of funding for all the consortia work. Seven Bridges could set up a billing group and limit billing group membership to individuals within the DCC. The DCC could set up all the necessary platform projects for the consortia using that one billing group.
This enables the DCC to use one platform billing group to track cloud costs for the whole consortium and manage invoicing. Alternatively, if the analysis working groups had their own sources of funding separate from the DCC, the projects could be configured to use different billing groups, with appropriate groups invoiced on a monthly basis.
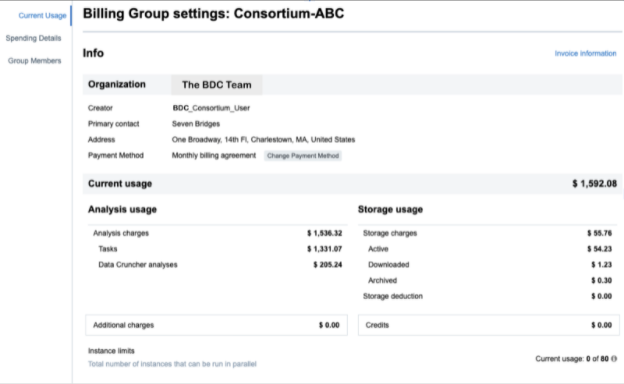
Figure 7. Billing groups track cumulative analysis and storage costs in addition to Task level expenditures. Shown here is the “Current Usage” information for a hypothetical “Consortium ABC.”
Learn more
- Cloud costs and payment (if you need any assistance to set up billing groups on BioData Catalyst, please contact [email protected]
- Estimate and Manage Your Cloud Costs.
Cloud Agnostic
The BioData Catalyst platform is cloud agnostic, and gives users the option to choose their preferred cloud location, on either Amazon Web Services (AWS) us-east-1 or Google Cloud Platform (GCP) us-west-1. In this way, users have the flexibility to store their raw data on either cloud provider and run the computation on the data where it lives.
For example, if a Study Coordinating Center has raw data stored in their own private AWS bucket, they could connect that cloud bucket directly to the platform.
Recommendation
In order to avoid egress costs, we recommend selecting AWS us-east-1 as the cloud location, if your data is stored in a AWS bucket in us-east-1. All the computation within the project will be performed on AWS us-east-1 and unnecessary egress costs between cloud providers can be avoided
In some cases, Study Coordinating Centers may have data stored in different locations (for example, one dataset on AWS and another dataset on GCP) because the universities involved may have agreements with different cloud providers.
Recommendation
In these situations, the data upload projects can be configured to the cloud location that matches where the Study Coordinating Center raw data is stored. When the Data Coordinating Center links all the datasets to one centralized project for harmonization, data egress is based on the cloud location of the Harmonization project.
In cases where the Study Centers all have their data stored locally, the DCC should decide on a cloud location for all of the platform projects as part of the setup process.
Recommendation
By using the same cloud location for all consortium projects, data egress costs between cloud locations will be avoided as data is linked between projects for sharing purposes.
Learn more
- To learn more about how the BioData Catalyst platform and Seven Bridges platforms solve this distributed data problem, please see our related blog post.
- Choose a cloud location for a project during project creation.
In summary
By keeping the above recommendations in mind, consortia members will find that the BioData Catalyst platform platform greatly simplifies and streamlines collaboration on a large scale.
If you have any additional questions on any topics covered in this guide, please contact [email protected] and the team will assist you as soon as they are able.
Updated 10 days ago
