Share content through Public Projects
Overview
BDCatalyst users have the opportunity to broadly share analysis examples, notebooks, tools/workflows, and data with the user community through Public Projects. Public Projects are platform projects that are accessible to all users on the platform from the top navigation bar.
Reasons to make a public project
Public Projects provide a space for researchers to publish their analyses with open access sample data, detailed walkthroughs, and contact information for feedback and improvements. Both researchers developing new tools and researchers using preconfigured pipelines benefit from published Public Projects.
- Researchers may be required to share content by a funding agency or for publishing.
- Course instructors need a mechanism to distribute tutorial content to trainees.
- Other researchers may simply desire to make their methods accessible and reproducible. In kind, many researchers come to BDCatalyst to find preconfigured workflows or notebooks that can inform and expedite their data analyses.
Considerations
Before you request to make your project public, consider the following:
Eligibility
Consider the best venue for your content. For instance, if you built a new workflow, the Dockstore tool registry may be a better option. However, if you have a collection of new tools or notebooks with similar themes and wish to release those with a written guide and sample data, then the Public Project space might be the right place. The Public Project should feature a unique addition such as newly developed tools/notebooks that other researchers would want access to. You should also consider whether your Public Project could be used with open access data, or is only usable with controlled access data. To discuss whether Public Projects are best venue for your content, please contact [email protected] or the BDCatalyst Program Manager.
Files
Providing workflows or a collection of notebooks to the community creates access to methods, but including practice data creates reproducible materials other researchers can learn from and build on. Consider which data you used while developing the project and if it can be made public. The project should have a small volume (under 5 GB) of sample files to support ease of use. The data must be open access and have appropriate approvals for public release. For instance, TOPMed hosted data cannot be included, but 1000 Genomes data meets the open access criteria.
Documentation
In order to publish the project, you must include documentation about what the project is and how to use it. The Description field on the project Dashboard is the perfect place to include detailed information on the purpose, included data, and how a user should explore or use the project. This section allows for markdown so you can clearly structure what is included. We also recommend providing a link to your repository (e.g., github).
Support
Seven Bridges Support will provide initial communication for users who have questions about the project. However, we recommend providing contact information for users to reach out directly to the developer’s team in cases of specific technical issues. For instance, a user might discover package deficiencies in the Public Project notebooks.
The Seven Bridges team will not update projects without developer support. If Seven Bridges cannot reach a project developer, the Public Project might be removed from BDCatalyst.
Preparing your project
Title
The project title should be short and informative. For instance, “Image Analysis Tool” might describe what the project contains, but it does not provide a researcher with much insight as to how they should use it. A better option could be “COVID-19 Image Segmentation with Deep Learning” which tells other researchers the type of image analysis tools in the project and the context it was developed. For formatting, always capitalize the first word as well as all nouns, pronouns, verbs, adjectives, and adverbs. Articles, conjunctions, and prepositions should not be capitalized.
Description
The project description should be cleanly organized using markdown to delineate titles and subsections. This section should begin with the title and then provide a few paragraphs describing the project, its purpose, and the advantages of this analysis. Each tool, workflow, or notebook should be included here with a short summary of its use case. Additionally, please include a summary of sample files and the analyses they apply to. This section also should detail what type of compute instances are required and if network access is required. Instance type is especially important to include if GPUs are required.
The Description can include a troubleshooting guide that explains common issues and important notes regarding the analyses. Contact information for your research group or your git repository for user feedback is appropriate to include.
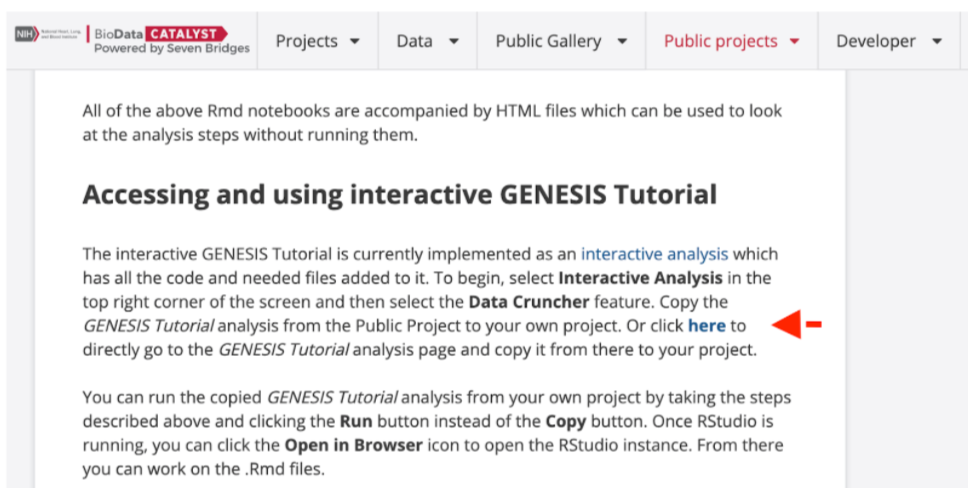
The end of the Description must include details on how to copy the Public Project to a private project as demonstrated in the GENESIS Tutorial.
Tools/workflows and tasks
All included tools and workflows should be wrapped according to best practices. Only the necessary and most recent tools should be included, and all should be descriptively named. For instance, if you include RNA STAR aligner, there should be one tool labeled “RNA-seq Alignment - STAR 2.7.3a” and duplicate previous versions such as “RNA-seq Alignment - STAR 2.5.4b” or “rnaalign_v1” should be removed.
The project should not contain any failed, aborted, or draft tasks. As you troubleshoot the project, you will accumulate a list of failed tasks. Before submitting the project to Seven Bridges for review and publishing, please clear all failed tasks. You should include a single set of completed tasks in the project as demonstration of successful use.
Notebooks
If notebooks for Interactive Analysis (R Studio, Jupyter Labs, SAS Studio) are included they need appropriate names similar to the tools and workflows. “Script_1” is not an appropriate name for publishing, but “Cloud Agnostic Data Import Script” is. In the notebooks, all cells should be executable, and no extraneous code should be included (i.e. if you have a “print” function for troubleshooting while developing the script, that should be removed before publishing). The beginning of the notebook should be appropriately annotated with the purpose of the notebook. Moreover, all modules and libraries that are needed should be loaded and commented at the beginning of the script. Throughout the script, please provide clear and detailed comments with consideration of users unfamiliar with the analysis.
Files
The project should only have files that are necessary to demonstrate the tools. There should be no duplicate or derived files. If you can use hosted open access data such as 1000 Genomes, that should be clearly noted in the Description and the files can be linked to in the project. Any non-hosted files should have clear titles: human_g1k_v37_decoy.fasta is descriptive enough, but test.txt is not. As much relevant metadata as possible should be included with each file including a description, the source of the file, if the file was modified (e.g., fasta is a subset of XYZ.fasta from ABC study), and any other relevant information.
Controlled data cannot be included with the Public Project. Any data that requires institutional approval or controlled access must be removed before submitting the project to Seven Bridges for publishing.
All projects should include a CHANGELOG file. This file must be updated with version information for the project (changed tools/workflows, added/removed files, changed notebooks, etc.). For instance, when “RNA-seq Alignment - STAR 2.5.4b” is updated to “RNA-seq Alignment - STAR 2.7.3a” then the changelog.txt should reflect this update. All additions to the CHANGELOG should include the date for the update.
Working with Seven Bridges
If you would like to share a Public Project, contact [email protected] with the following note:
Hello,
I have a project that I would like the Seven Bridges team to make into a Public Project. The purpose of this project is [to be filled in]. Can you please connect me with the Program Manager for BDCatalyst for next steps?
Sincerely
[Your name]
The Program Manager will set up a meeting to discuss eligibility and goals for the project. If Public Projects is the best place for publishing the work, then the Program Manager will connect the researchers to the Seven Bridges Bioinformatics team.
The Seven Bridges Bioinformatics team reviews and approves projects for publication. The research team should have a clean project following the guidelines outlined above. The Bioinformatics team member will be added to this project for review. The review includes clarity of the description, function of the tools/workflows/notebooks, and inclusion of appropriate files. Moreover, this private project can be used as a pre-publishing draft project for future updates.
Once the Program Manager and Bioinformatics team approve of the project, it will be copied to a new Public Project and released on BDCatalyst.
Future Updates
This initial release of an analysis pipeline is rarely the final release. After your project has been published under “Public Projects” on Seven Bridges, it is likely you will want to update the project based on updated tools, found bugs, or user feedback. This process should be planned before the project is published. Currently, users cannot update projects directly. However, a researcher can add a Seven Bridges Bioinformatics team member to a pre-published private project. This project can be used for final updates that the user-research team wants added to the platform.
Updated 8 days ago
