TOPMed Annotation Explorer
The TOPMed Annotation Explorer enables you to interactively explore, query, and study characteristics of an inventory of annotations for the variants called in TOPMed studies.
This application can be used pre-association testing, to interactively explore aggregation and filtering strategies for variants based on annotations, and generate input files to use for multiple-variant association testing.
It can also be used post-association testing to explore annotations associated with set variants, say for example, variant sets found significant during association testing.
Prerequisites
- API token - to be able to use the Annotation Explorer, you will need your Platform authentication token. Learn more on how to generate the token.
- Access to TOPMed data - which is approved and managed by dbGAP; researchers can still try the Annotation Explorer with the open access Freeze5 variants (see below).
Available datasets
The Annotation Explorer currently hosts a subset of genomic annotations obtained using Whole Genome Sequence Annotator software for TOPMed variants. Currently, annotations for TOPMed Freeze8 variants and TOPMed Freeze5 variants are integrated with the Annotation Explorer.
Researchers who are approved to access one or more of the TOPMed studies with Freeze8 variant data will have the ability to access the Freeze8 annotation data on the Annotation Explorer.
Similarly, researchers who are approved to access one or more of the TOPMed studies with Freeze5 variant data will have the ability to access the Freeze5 annotation data on the Annotation Explorer.
Researchers without access to TOPMed studies can try out the Annotation Explorer with the open access Freeze5 variants submitted to dbSNP or the open access Genome_wide annotations for all combinations of SNVs, including each position in the genome as well as INDELs submitted to dbSNP.
IMPORTANT BILLING NOTES
Daily quota for querying is set to 6TB. Daily quotas reset at midnight Pacific Time.
Copy Annotation Explorer from the Public Project
The Annotation Explorer is currently implemented as an R shiny application. Users launch the application as an interactive analysis which has all the code to launch an interactive R shinny application.
The first step is copying the Annotation Explorer public project.
Procedure
- Click Public projects in the main navigation and choose Annotation Explorer.
- Open the **Data Studio" tab.
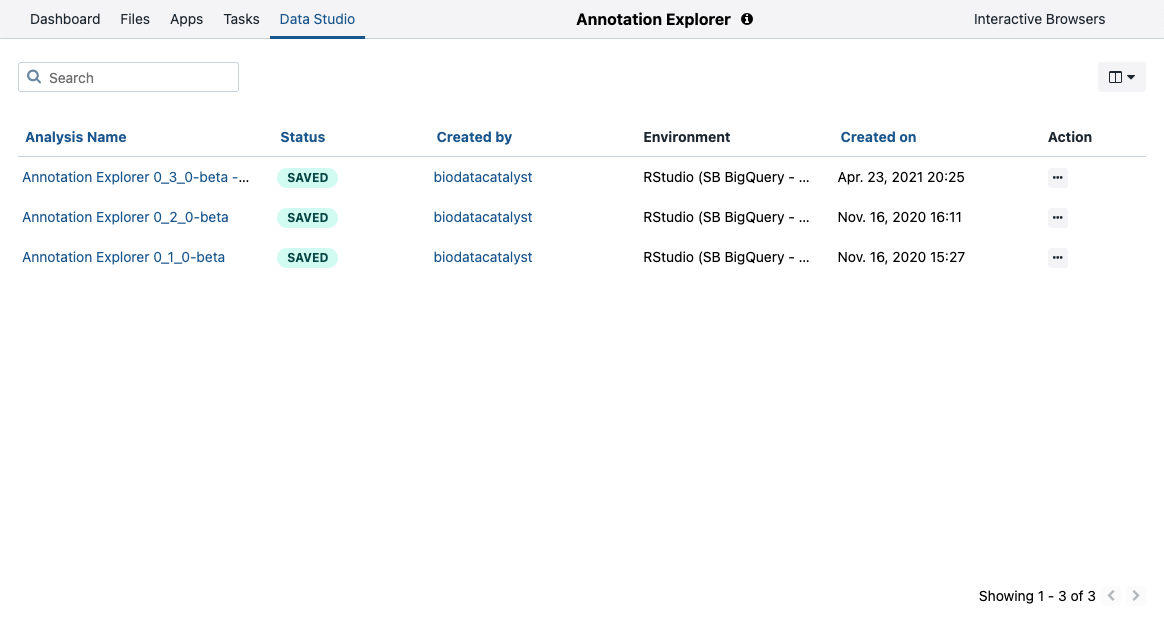
- Under Action click and select Copy.
- Name your analysis and choose the project you want to copy the Annotation Explorer to.
- Click Copy.
The Annotation Explorer is now copied to your project. The next step is launching the Annotation Explorer.
Several versions of the Annotation Explorer analysis are available and the latest version will have the Latest suffix in the analysis name.
Launch Annotation Explorer
Procedure
- Access the project you have copied the Annotation Explorer to.
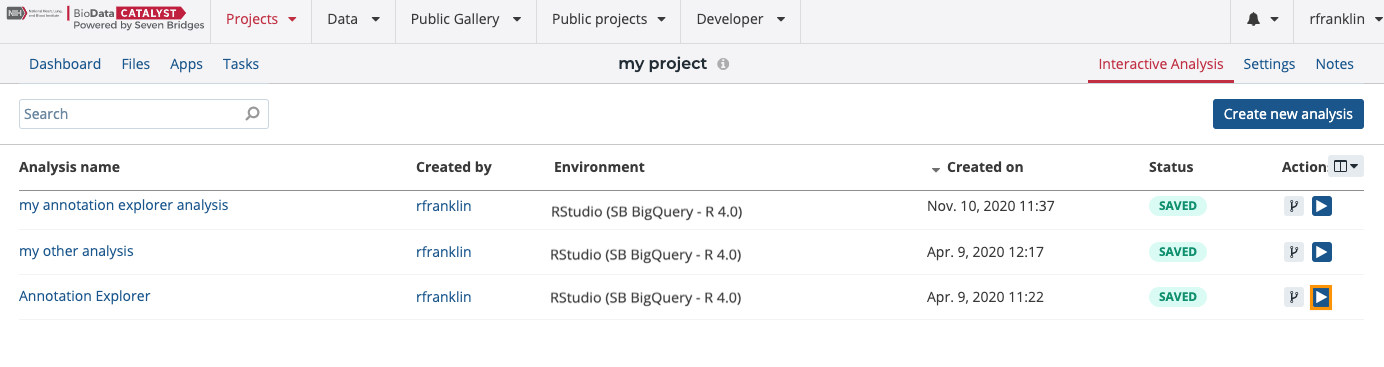
- Click Interactive Analysis in the upper right corner.
- Open the Data Studio.
- Click the run icon next to the Annotation Explorer analysis. The analysis will initialize within a sequence of 3 steps. You can monitor the progress.
- Once the RStudio instance is initialized and the status is changed to "Running", click the open icon on the far right.
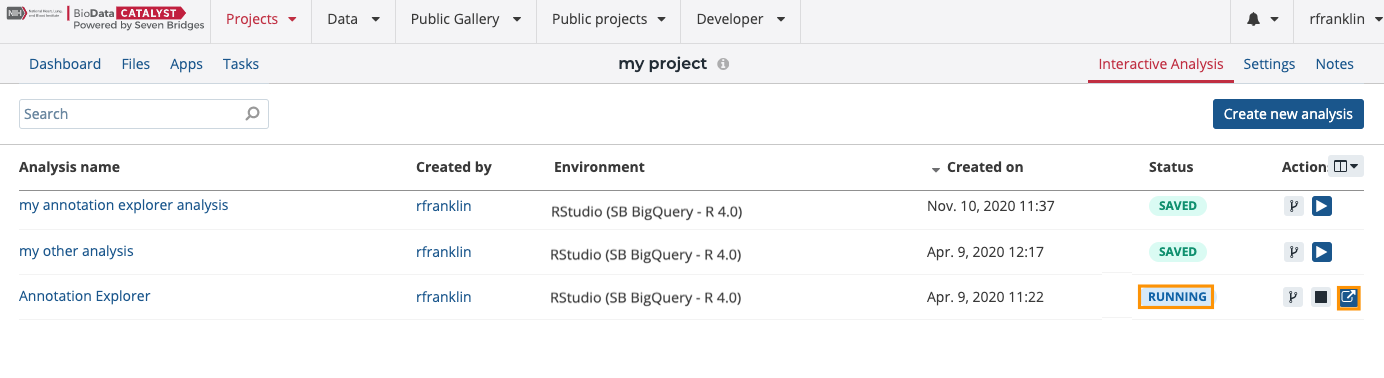
This will launch the R studio interface in a new tab of your browser with the app.R script.
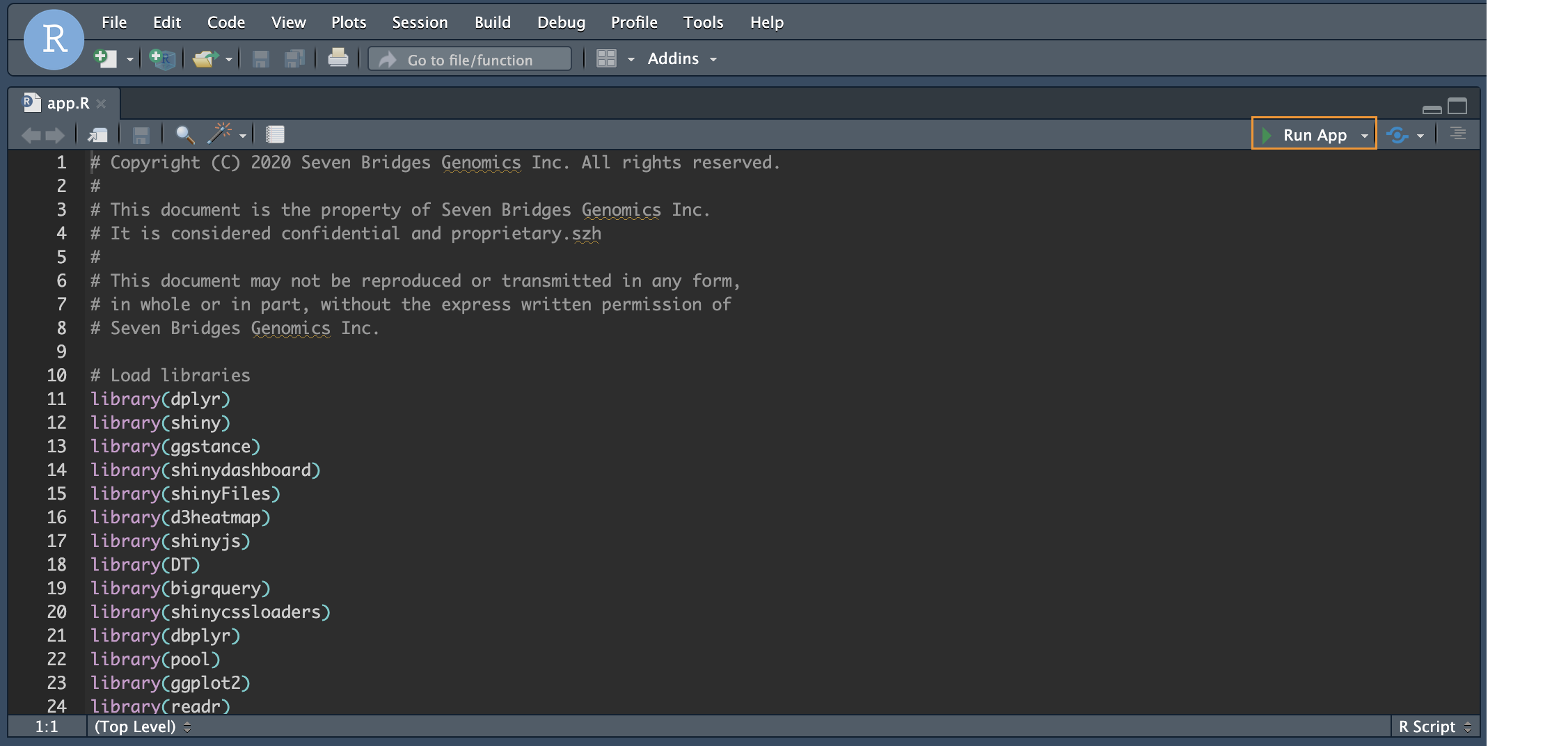
- Click Run App at the top right hand corner of the app.R script to launch the interactive R shiny app in a new window.
- Click Start Exploring to launch the Annotation Explorer.
- Follow the instructions to authenticate using previously generated Platform authentication token.
Rate limiting
The number of users that can use the Annotation Explorer at any given time is limited. If you are unable to access it, please try again after a few hours.
Once you start using it, your session will be limited to 3 hours and you will be allowed to use it again in 24 hours.
Updated almost 4 years ago
