Worked example of uploading SamTools Sort
See a video of this tutorial below.
Introduction
The CGC Software Development Kit (Rabix) allows you to add your tools to the CGC and use them to run analyses, as you do with tools that are already publicly available on the CGC. This is done by installing the tools in a Docker container and then describing their behavior on the CGC.
The first part of the procedure in deploying your tool to the CGC is to create a Docker container and install the tool in it. Once this is done, you need create a snapshot of the container, called an image, and push it to the CGC image registry (cgc-images.sbgenomics.com) or the official Docker image registry, Docker Hub.
The second part of the procedure is to specify the tool's behavior on the CGC, including its inputs and outputs, runtime requirements, and execution semantics. The specification is entered using the Tool Editor. This allows you to use the tool as an individual application on the CGC or interface it with other tools and create workflows.
Objective
This tutorial demonstrates how to install and describe the sort subcommand of SamTools on the CGC. Specifically, we shall: install the bioinformatics package SamTools into a Docker container, push it to the CGC image hub, and describe the sort subcommand in the tool editor.
Prerequisites
For this tutorial, you will need:
- A CGC account.
- One of the following machines:
- A Linux computer with Docker installed on it. Full installation instructions are available here.
- A Mac with Docker Machine installed on it. Full installation instructions for OS X are available here.
- A Windows computer with Docker for Windows or Docker Toolbox depending on the Windows version Full installation instructions for Windows are available here.
On this page:
1. Create a project
2. Upload SamTools in a Docker image
3. Describe each subcommand tool in the graphical editor
1. Create a project
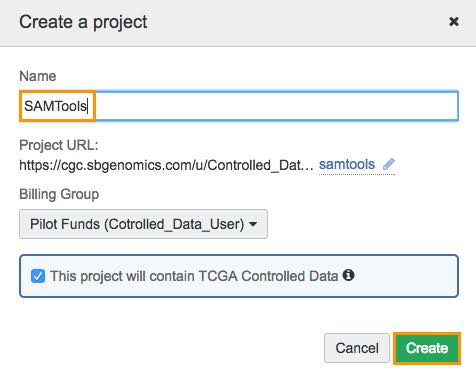
- Log in to your CGC account, and click Create a project in the main navigation bar.
- Name the project (e.g. 'SamTools', you can always delete this project when you've finished the tutorial.)
2. Upload SamTools in a Docker image
We will first use Docker image to create an image containing Samtools. We'll start with an Ubuntu base image, install SamTools, then commit and push the image to the CGC image registry, and push it to upload the tool to the CGC. This is illustrated in the example below. The username used in the example is rfranklin, the developer project name is samtools and the image tag is v1.
In the example, most of the steps taken are in order to get the tools needed to compile SamTools. These details will vary for different tools. As such, the example here should not be taken as a template for all command line tools.
To create a Docker image:
Uploading Docker images
If you haven't already seen it, take a look at the documentation on uploading Docker images.
- Open up a terminal to get started.
Depending on your operating system, first make sure that Docker is started:
- Docker on Mac OS 10.10.3 Yosemite or newer run Docker for Mac and start a terminal of your choice.
- Docker on Mac OS 10.8 Mountain Lion or newer run Docker Machine, by opening Docker Quickstart terminal or by using the command docker-machine start default.
- Windows 7 or 8: run Docker Quickstart Terminal.
- Windows 10: run Docker for Windows and start a terminal of your choice.
- Linux: skip this step.
- To install SAMtools in a Docker container, we will enter the following commands:
2.1 Log in to the CGC image registry (cgc-images.sbgenomics.com) from the terminal:
Docker login
Note that you should enter your authentication token in response to the password prompt, not your CGC password.
$ docker login cgc-images.sbgenomics.com # You should enter your authentication token in response to the password prompt, not your CGC password.
Username: rfranklin
Password:
Email: [email protected]
2.2 Load up a container from the Ubuntu base image, update the packages inside the container and install SAMTools:
$ docker run -ti ubuntu # Load up a container with the ubuntu base image and run bash inside
Creating container from image ubuntu
root@container$ apt-get update # Update the package index inside the container
root@container$ apt-get install wget build-essential zlib1g-dev libncurses5-dev # Install the tools we need to compile SamTools
root@container$ wget https://github.com/samtools/samtools/releases/download/1.2/samtools-1.2.tar.bz2 # Download the Samtools source code
root@container$ tar jxf samtools-1.2.tar.bz2 # Unpack the archive
root@container$ cd samtools-1.2 # Go into the directory containing the unpacked Samtools source code
root@container$ make # Compile the code
root@container$ make install # Install the resulting binaries
root@container$ samtools --version # Check that SamTools has installed
root@container$ exit
You can choose any Docker base image for your tool.
A Docker container is a running instance of a Docker image. Once you have instantiated a container from an image using the docker run command, the initial part of the command line will change to something in the form root@container, e.g. root@afa7af5b5d8b. The root part denotes that you are the root user within the container, while the part after the '@' symbol is the ID of the container. Once you have exited the container, copy the container ID as you will need it to perform the next step.
2.3 Commit the container to the image:
$ docker commit 19d574537671 cgc-images.sbgenomics.com/rfranklin/samtools:v1 # Grab the container ID '19d574537671' from the command prompt inside the container you just exited, to commit its image
7f7f2b36bffd5dae5d8e4c699079aa96379f5075ce175fb4abd0197a46ebfcd3
The repository name used in this example is rfranklin/samtools, following the <user_name>/<project_name> pattern. Please note that the allowed characters for repository names are lowercase and uppercase letters, numbers 0 to 9, dash (-) and underscore (_). Learn more about repositories in the CGC image registry.
2.4 Push the image to the CGC image registry:
$ docker push cgc-images.sbgenomics.com/rfranklin/samtools:v1
The push refers to a repository [cgc-images.sbgenomics.com/hodesdon/new] (len: 1)
...
latest: digest: sha256:d2304a53961b9e8215805448d0738a0174b3b18ee6ea6145bf1d0062d615ae1a size: 8039
3. Describe each subcommand tool in the graphical editor
We have created a Docker image with SamTools inside, and pushed it to the CGC image repository. To use SamTools on the CGC, we still need to capture its interface with the tool editor, so that it can be integrated with other CGC tools.
The tool editor treats each subcommand of a command line tool as a distinct tool. So, in this example, we will just describe the SamTools subcommand sort.
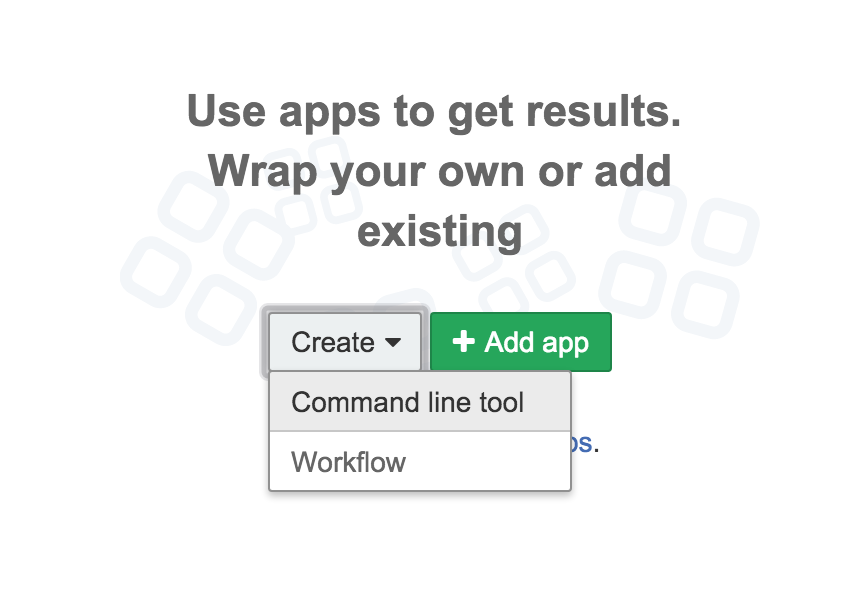
- The tool editor can be accessed from inside the project that you created on the CGC. Go to the dashboard for the project, and click 'Create' on the panel marked 'Apps'. This brings up a drop-down box. Select Command line tool to describe a new tool.

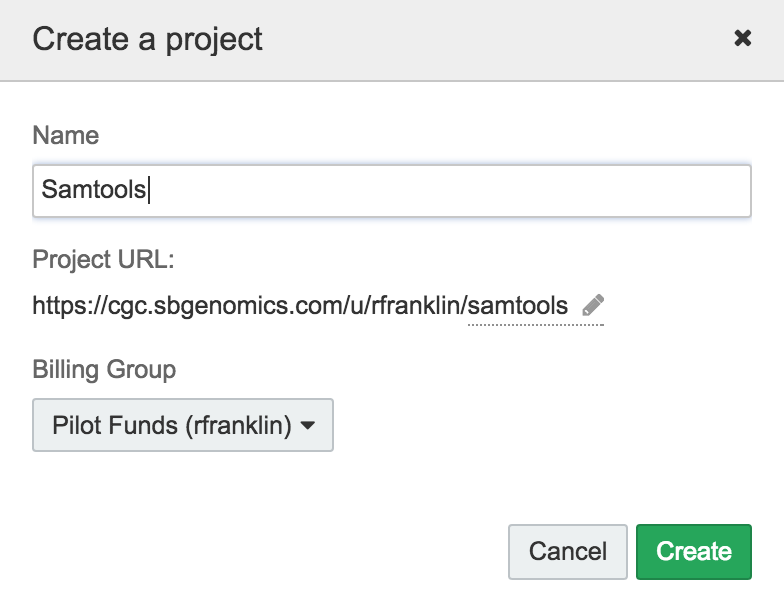
- To add the tool, first give it a name. Let's name the SamTools
sortsubcommand 'SamTools-sort':
- Once you have given your tool a name, and clicked Create, the graphical tool editor interface will open. This contains fields with which you can characterize the format of the subcommand as it is executed on the command line.
Since we're going to characterize the sort subcommand, let's take another look at its usage. We can use sort by entering samtools sort on the command line, inside the Docker container where we installed SamTools.
Since we exited the container in which we installed SamTools, we need to open it up again so that we can query the usage of SamTools sort. Do this using the docker run command again, but this time specifying the image from step 2:
$ docker run -it cgc-images.sbgenomics.com/rfranklin/samtools:v1
Rabix CLI
If you're using the Rabix CLI use the command
cgc docker-run <image>where< image>is either the image ID or<repository>/<tag>.
Now let's check the usage of the sort subcommand:
$ samtools sort
Usage: samtools sort [options...] [in.bam]
Options:
-l INT Set compression level, from 0 (uncompressed) to 9 (best)
-m INT Set maximum memory per thread; suffix K/M/G recognized [768M]
-n Sort by read name
-o FILE Write final output to FILE rather than standard output
-O FORMAT Write output as FORMAT ('sam'/'bam'/'cram') (either -O or
-T PREFIX Write temporary files to PREFIX.nnnn.bam -T is required)
-@ INT Set number of sorting and compression threads [1]
We can see that the sort subcommand takes the options listed above, followed by an input file, in.bam.
Let's suppose we want sort to output a file named 'output.bam'. In this case, given the usage of sort we need to use the following options:
- The default behavior of the subcommand is to write the sorted output file to standard output. However, this default behavior can be overridden to instead produce output as a file named 'output.bam', using the option
-o output.bam. - Since we want to output a BAM file, we need to specify the file format as well as the filename. We do this with the option
-O bam. - The
-Toption says it is required. This fixes the prefix of the temporary files. Let's prefix our temporary files withtmp_. We do this with the option-T tmp_.
To achieve this behavior for an input file named 'unsorted.bam', we would need to execute the following command: samtools sort -O bam -T tmp_ -o output.bam unsorted.bam
We will describe our required behavior in the tool editor by entering the information specific below:
See the documentation on the Tool Editor for more information on the fields below.
Within an application, there are five tabs, General, Inputs, Outputs, Additional Information, and Test. We will walk through the steps required to describe a tool in each of these below.
General Tab
Docker Repository[:Tag]: This is the location of the image containing the command. Its format is images.sbgenomics.com/<repository><:tag>. In our example we would enter: cgc-images.sbgenomics.com/rfranklin/samtools:v1
CPU: We'll leave this with the default of 1.
Memory: Set the value to 5000 MB of RAM. This amount of memory is needed to process the BAM file that will be used as the input.
Base Command: The base command is the part of the command that precedes any arguments; in other words, it is the command and subcommand, if there is one. In our example, samtools-sort, we enter samtools sort into this field. The editor splits base commands on spaces, so this entry will split into a field containing samtools and a field containing sort.
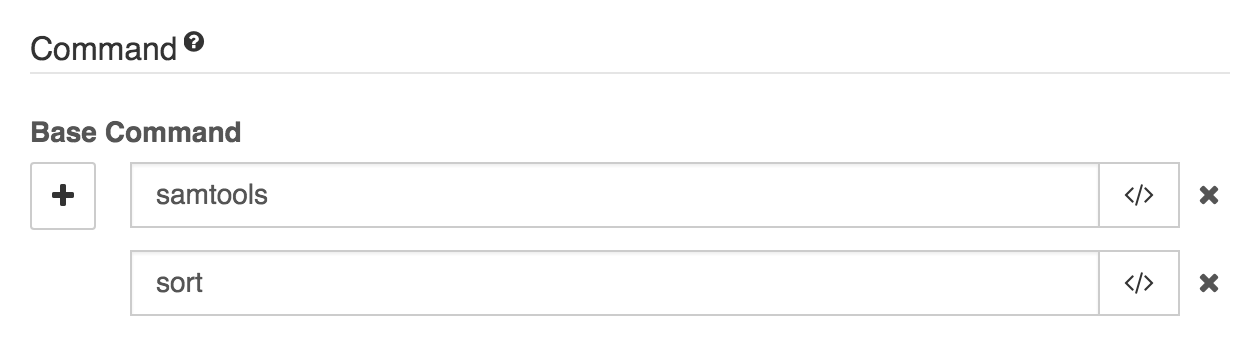
Under Command, enter 'samtools sort' and the GUI will break this into the base command 'samtools' and the subcommand 'sort'.
Stdin, Stdout: We can leave these blank. We decided to pipe output to a file instead of standard output.
Success code and Temporary fail code: Set these to 0 and 1 respectively. This is standard behavior.
Arguments: We want SamTools sort to output a BAM file named 'output.bam'. As described above, we can make it do this using the following code: samtools sort -O bam -T tmp_ -o output.bam unsorted.bam
We can enter the arguments for this command in the Arguments field of the tool editor, as follows:

Click on the '+' to being entering arguments for the command line.
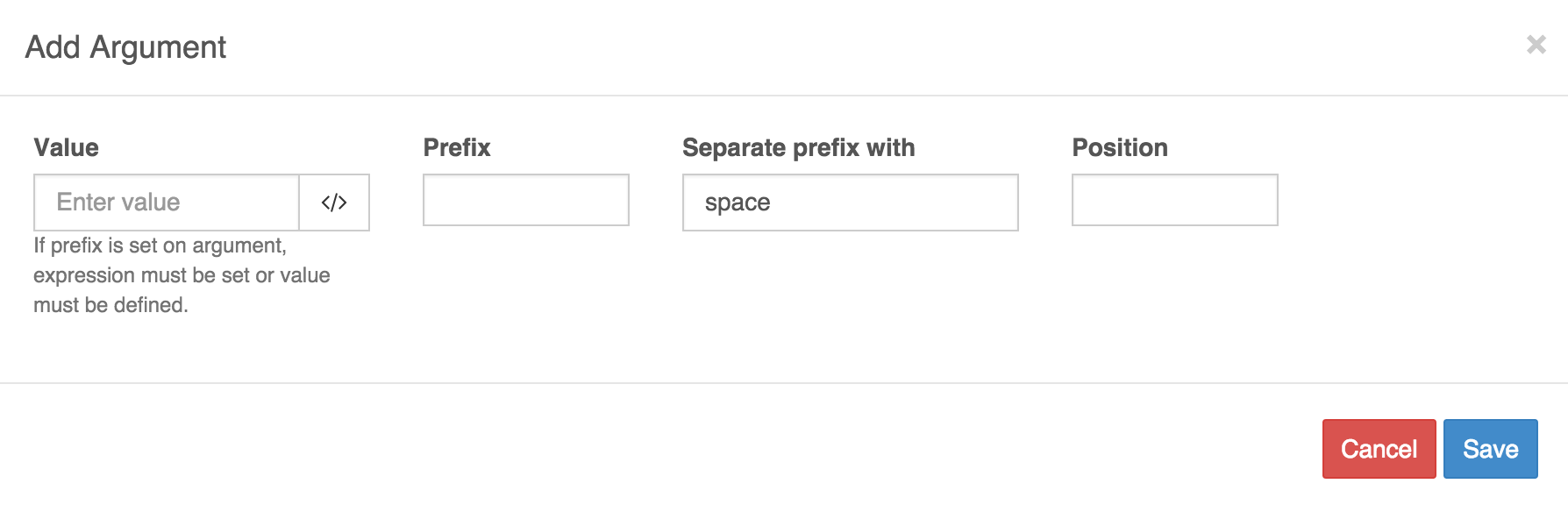
A dialog window will pop up with default fields.
Argument 1:
- Value:
bam - Prefix:
-O - Separate prefix with:
space - Position:
1
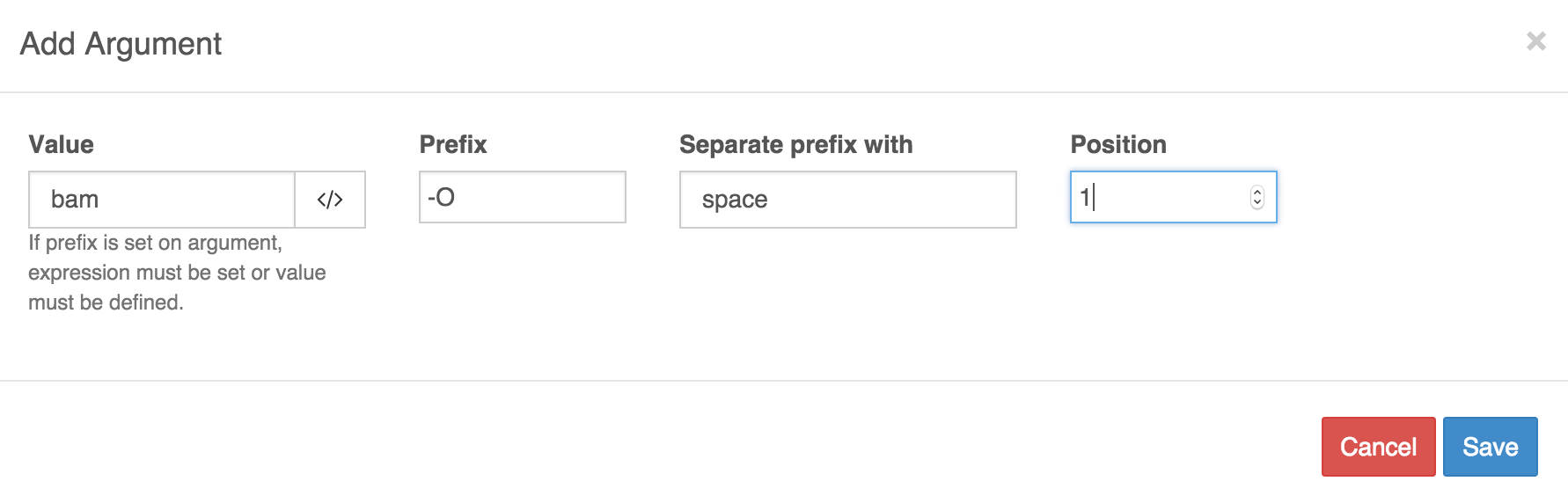
Enter the appropriate values and Save.
Argument 2:
- Value:
tmp_ - Prefix:
-T - Separate prefix with:
space - Position:
2
Argument 3:
- Value:
output.bam - Prefix:
-o - Separate prefix with:
space - Position:
3
These resulting Arguments settings are shown in the following screenshot:
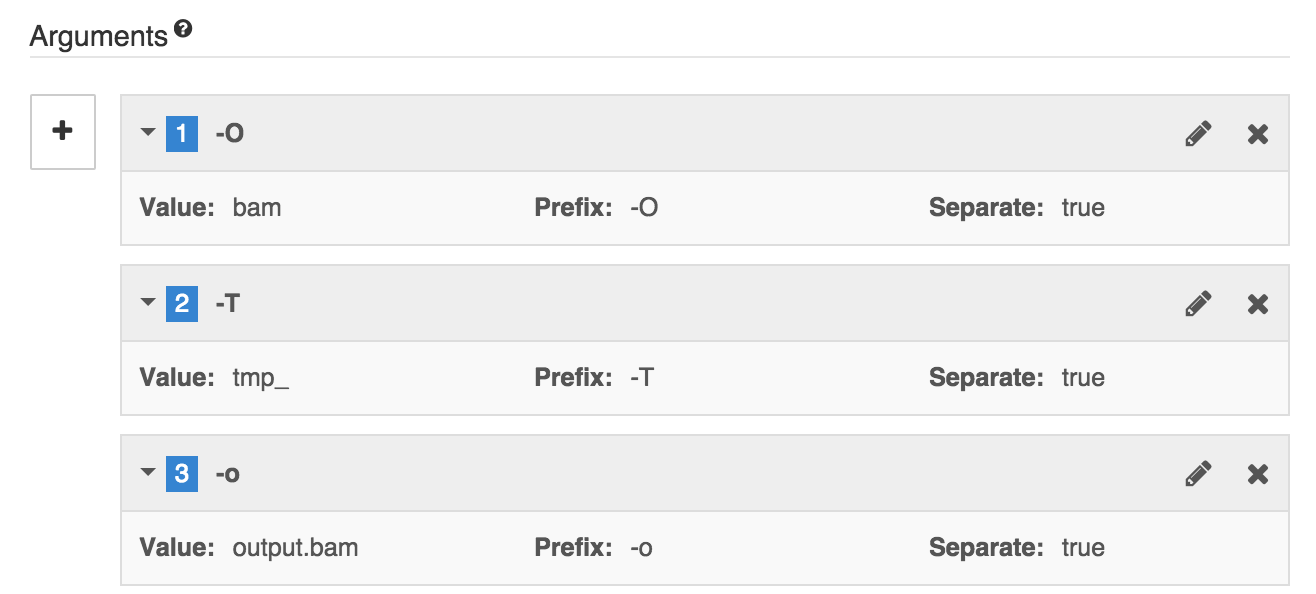
Once done, this is what you should see in the Arguments section.
When you have finished, the General Information tab should look like this:
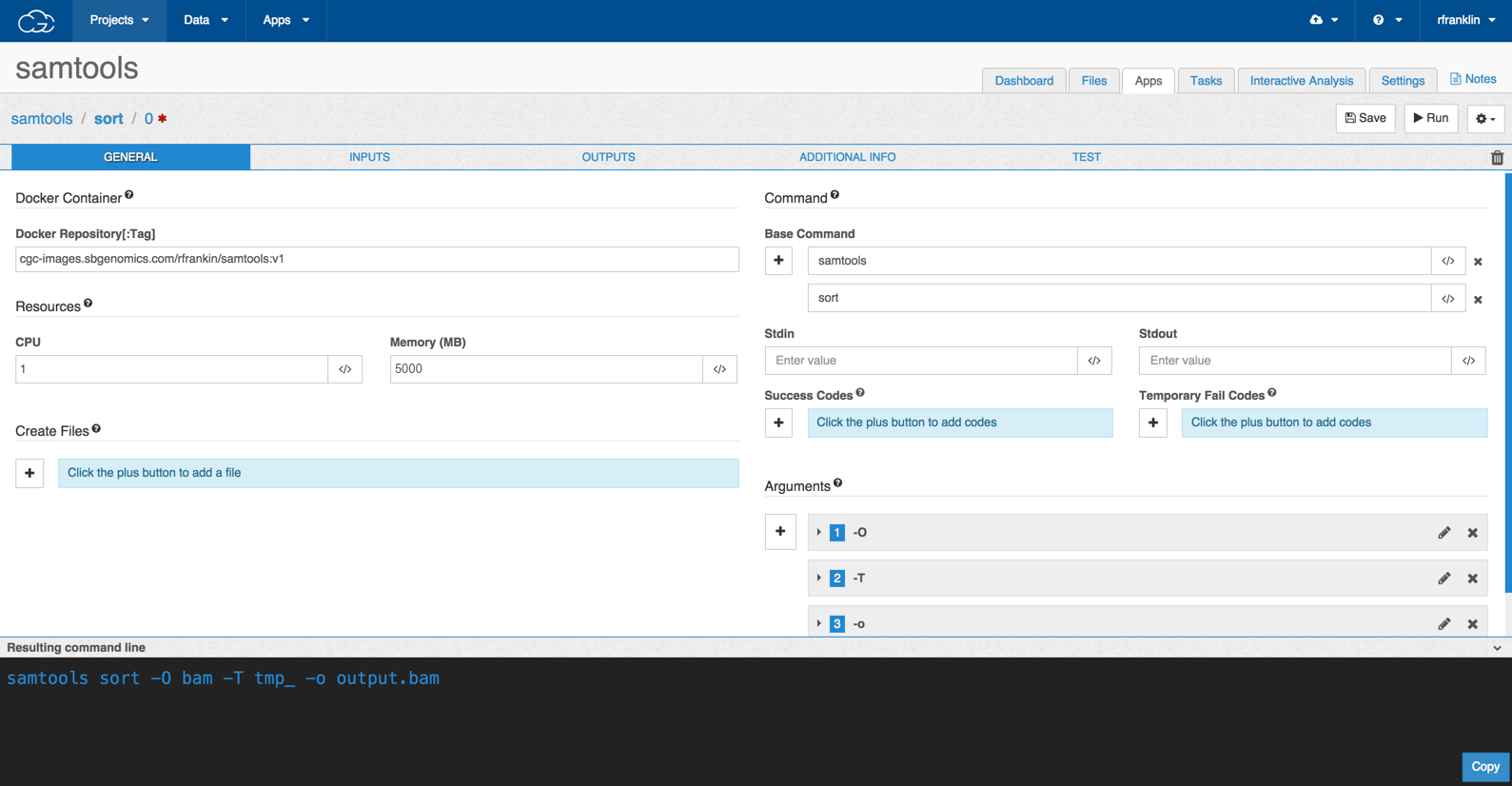
The Inputs Tab
- Click the + button to add an input port.
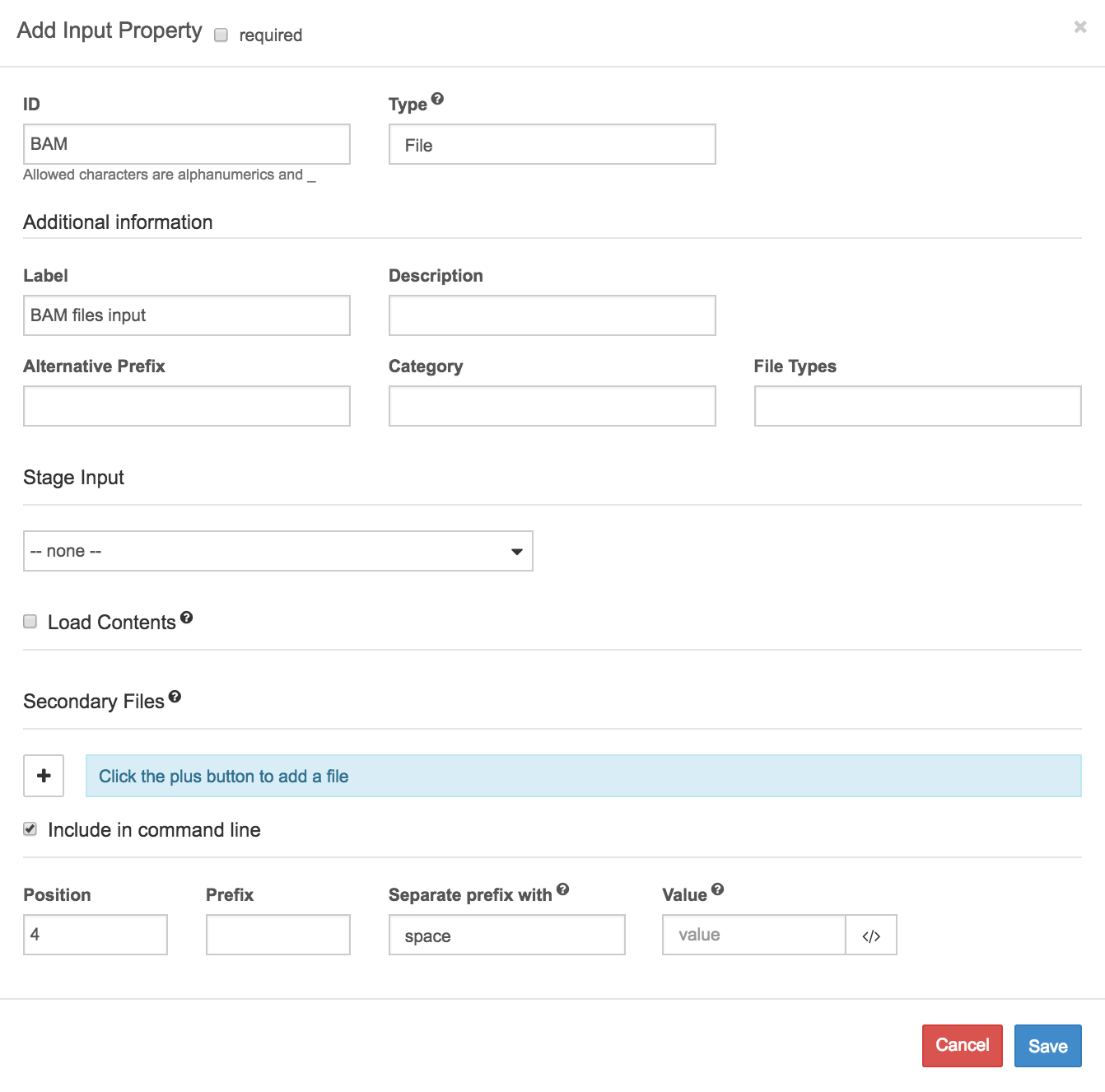
- Set the ID of the input to 'BAM' to label it as the port where BAM files are inputted to the tool. Set its Type to 'File'.
- Enter a Label to be displayed on graphical interfaces: we went with 'Bam files input'. Enter a description as well, if you like.
- There are no Secondary files so we can leave this blank.
- Check the box marked Include in command line to enable command line binding. This indicates that when the file is executed on the command line, file inputs are entered into the terminal on the command line.
- Under the checkbox to Include in command line, we enter the details of how inputs are entered to the command line.
a. Leave the Value field: we'll enter files directly on the command line.
b. Leave the Prefix field empty as well. This indicates that files are entered to the terminal with no preceding option to indicate the input (although there may be preceding options to control other aspects of the command).
c. Set the Position to 4, to indicate that the input file comes after the-O bamoption, whose position we set to 1, the-T tmp_option, whose position we set to 2, and the-o output.bamoption, whose position we set to 3.
d. This setting indicates that for an input file named unsorted.bam passed to the command line tool, the full command will have the form:samtools sort -O bam -T tmp_ -o output.bam unsorted.bam
The Outputs Tab
- Click the + button to add an output port.
- Set the ID to name your output port. We'll name this one 'sorted'.
- Set the Type to 'File' for you sorted files.
- Set the glob field to '*.bam'. This use of globbging will pattern-match any file that ends with '.bam' and report is as the output of the tool.
- Enter a label for the port, which will be used on any visual interface the tool appears in, such as the workflow editor.
- Specify the File Types that the port produces, in this case BAM files.
- In this example, we haven't annotated the output files with metadata or included secondary files (like index files) so we can leave the rest of the fields blank.
The Test Tab
- Fill in some dummy input values of the kind you would enter as command line arguments to the tool. Then you can inspect the resulting command line, at the bottom of the tab.
- The tool has a single input port for files, so we can enter a BAM file name, 'unsorted.bam' as a dummy input for this port. Notice that the command line output at the bottom of the screen changes to show the command that would be executed on the command line if a BAM file named 'unsorted.bam' were stipulated as the input file for the SamTools sort subcommand. The resulting command in this case is:
samtools sort -O bam -T tmp_ -o output.bam unsorted.bam. This is what we'd want to see in order to sort 'unsorted.bam'. So, our tool description looks like it was successful!
The Additional information Tab
Here you may enter some details about SamTools sort to give more information about the tool's developers and uses. All the fields on this tab are optional.
When you've finished, click Save.
That's it! SamTools has been Dockerized, and its sort subcommand can now be executed on the CGC at the touch of a button, either on its own or as part of a workflow. To run SamTools sort we just need to add TCGA data to a project. Then, we can click Run on SamTools sort, input the file, and obtain the results.
Video tutorial
Updated less than a minute ago
